I have seen that the "PQS Protein Quaternary Structure Query Form at the Protein databank in Europe - PDBe" has stopped working. (http://www.ebi.ac.uk/pdbe/pqs/). Do you know if these data are provided using another website?
Thank you;
I have seen that the "PQS Protein Quaternary Structure Query Form at the Protein databank in Europe - PDBe" has stopped working. (http://www.ebi.ac.uk/pdbe/pqs/). Do you know if these data are provided using another website?
Thank you;
In the old days, only the asymmetric unit was stored in the PDB. Now, I think all PDB structures also have the biological assembly associated with them, and it's the default shown on the PDB pages.
More Info @ PDB: Looking at Structures: Introduction to Biological Assemblies and the PDB Archive
FTP site: ftp://ftp.wwpdb.org/pub/pdb/data/biounit/coordinates/divided/
(This PQS page says: "The PQS service will not be incremented with new data from August 2009 and the service will be discontinued at some future date." which I think is further proof that the PDB superseded PQS.)
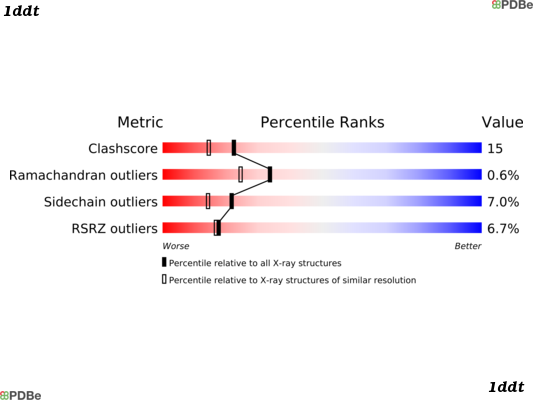
PQS is deprecated in 2009 and quaternary data is now available via PISA interface of PDBe Here is an example using 1DDT:
Monomeric structure of 1DDT

Homodimeric assembly of 1DDT via PDBe
You can download the 3D coordinates here
PISA would be the 'successor' of PQS I'd say. Also, as mentioned, REMARK 350, or BIOMT, can give you the matrices and translation vectors necessary to produce the biological unit from its separate entities.
Can PiQSi help?
Another resource that can help in elucidating the quaternary structure is EPPIC. It enumerates all interfaces present in the crystal and classifies them into biologically relevant or crystal contact by analysing the selection pressure patterns at the interfaces via multiple sequence alignments of sequence homologs to the protein in question. By doing this evolutionary analysis it offers a complementary view to PISA which estimates the binding energies from the contacts but does not look at sequences at all.
On the downside EPPIC does not offer the full quaternary structure prediction but only a list of interfaces that lead to the formation of the biological assembly. One needs to do some manual work to infer the quaternary structure from those.
Try this one
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.
thank you......