I am working with a program that generates artificial FASTQ files from a given reference genome and it allows for read length customization. I am trying to produce several FASTQ files with different read lengths. Hence, I want to know what read lengths are produced by real sequencers such as those from Illumina. I found this site which states the maximum read length of some sequencers.
However, when stating, for example, that the maximum read length is 2 x 150 bp, is that supposed to mean that each end of a contig will be 150 bp long? Also, if that is the case, does this mean that the sequencer could be set up to produce read lengths equal to any number of base pairs less than 150? i.e. could the sequencer produce read lengths of 2 x 1 bp, 2 x 2 bp, 2 x 3 bp,...., 2 x 148 bp, 2 x 149 bp? Lastly, which one of those sequencers is the most commonly used for genomes which are about as long as, say, that of E. Coli?

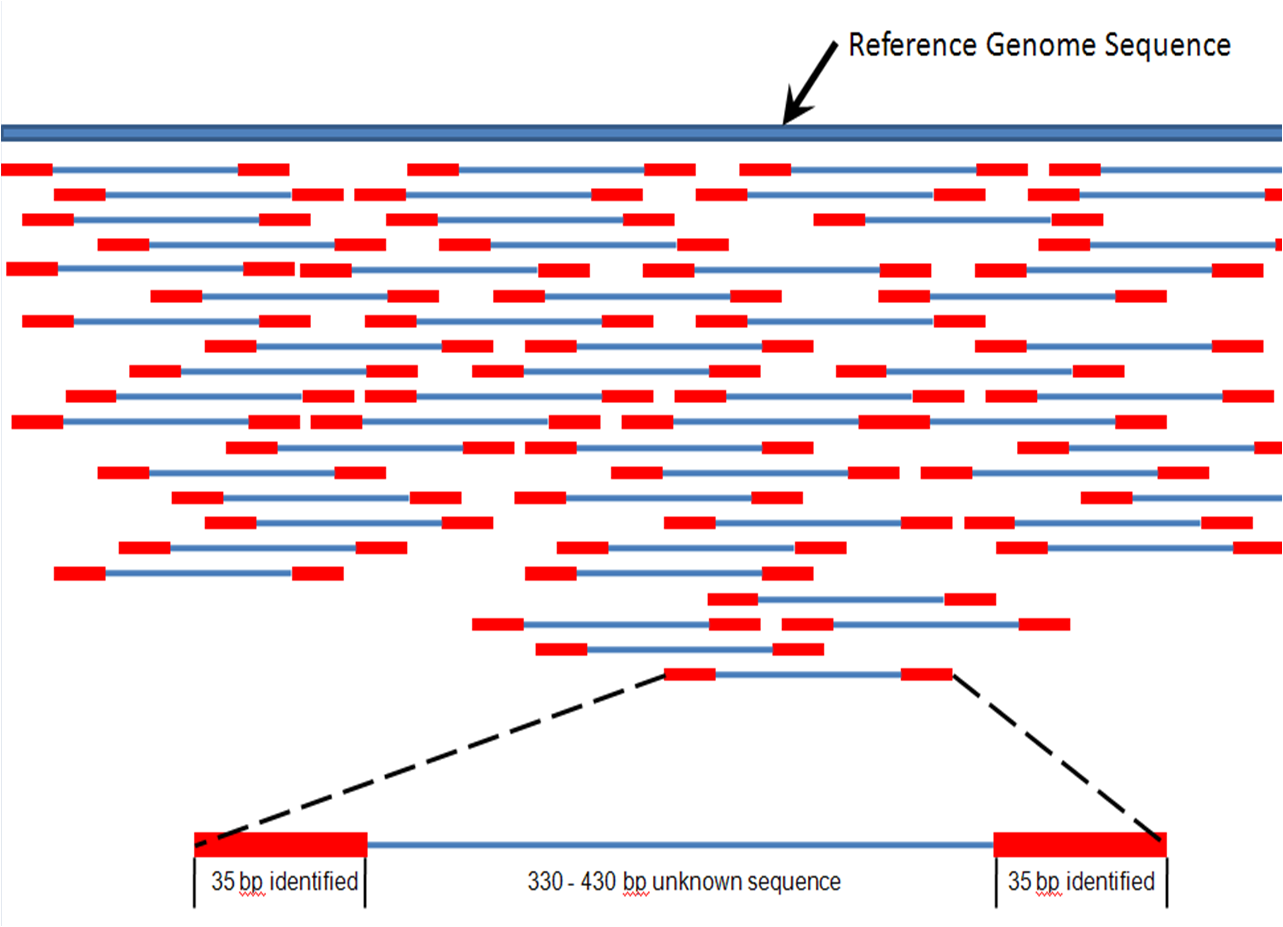

You can also take a look at this link. Best explanation of frequent terminologies - library (contig), insert, fragments, etc.