Every element of BLOSUM matrix is computed by the formula (from Wikipedia):

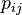 is the probability of two amino acids
is the probability of two amino acids 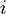 and
and  replacing each other in a homologous sequence, and
replacing each other in a homologous sequence, and 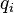 and
and  are the background probabilities of finding the amino acids
are the background probabilities of finding the amino acids 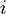 and
and  in any protein sequence. The factor
in any protein sequence. The factor 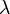 is a scaling factor.
is a scaling factor.
I would like to compute probability of replacing one amino acid 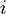 by another
by another  . The BLOSUM matrix is implemented in biopython module, but unfortunately I have not found the probabilities
. The BLOSUM matrix is implemented in biopython module, but unfortunately I have not found the probabilities 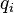 and
and  . Is there any easy way to obtain it or compute it?
. Is there any easy way to obtain it or compute it?


Please upvote and "accept" if this answered your question.