I am wanting to visualize a tree with more than 50,000 OTUs and save it as an image. Which software would be suitable? I have tried MEGA but it failed at this point. It seemed that FigTree also worked slowly since the tree is so big.
Which software can be used to visualize phylogenetic trees with a large number of OTUs (e.g. 50,000)
3
Entering edit mode
10.4 years ago
Sishuo Wang
▴
230
4
Entering edit mode
10.4 years ago
jhc
★
3.0k
The ETE toolkit might help you. You have different options:
- Collapse the tree before drawing, so the shown tree is easy to handle. Note that ETE will allow you to search and for target nodes, collapse them and save information about the content of each collapsed branch. The ETE GUI will allow you to click and inspect your node annotations)
- Split the tree in parts, so you can inspect each of them separately
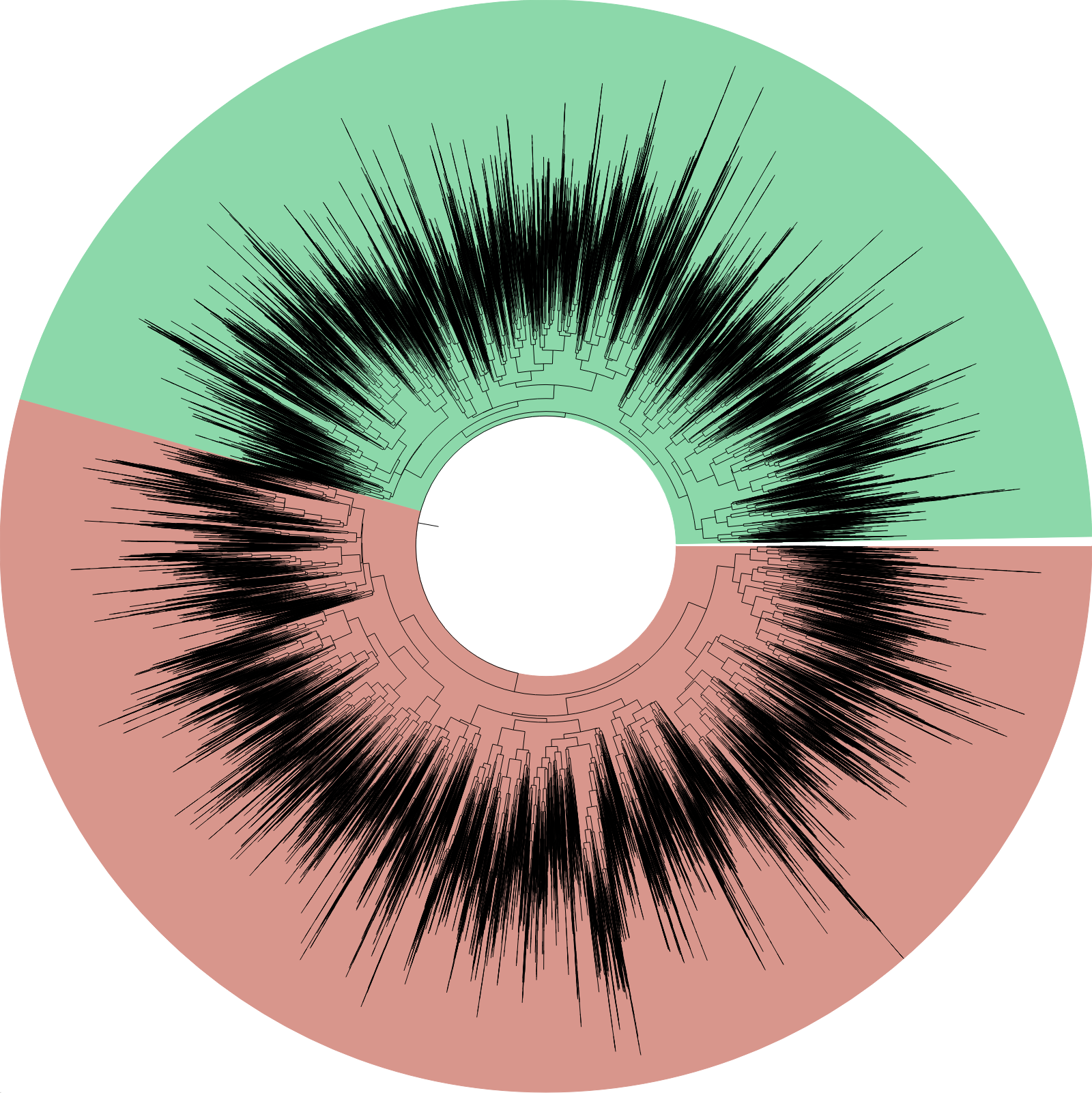
- Draw the whole tree as a PNG image: Only the general structure of the tree will be recognisable, so it makes no sense to print text or very elaborate information in the image. However, ETE will be perfectly capable of producing images for trees this large and you can always play with color lines, background, collapsed regions, branch scale, etc... so your figure is somehow readable and fits your purposes. I attach a simple img generation script that I have tested with a random tree of 50,000 tips. It took 60 secs and 5G of RAM in an average linux machine, and generated this output:
None of this options will allow you to browse the tree interactively and as a whole. This is a main line of development in ETE, but currently I only have alpha versions of the software to do this type of things.
import sys
from ete2 import Tree, TreeStyle, random_color
def clean_ly(node):
node.img_style['size'] = 0
t = Tree()
t.populate(int(sys.argv[1]), random_branches=True)
ts = TreeStyle()
# set a clean view for the tree
ts.show_leaf_name = False
ts.mode = 'c'
ts.layout_fn = clean_ly
# Drawing text will be useless in such a big trees. Perhaps colors indicating
# different regions of the tree
c1 = random_color(l=0.7)
t.children[0].img_style['bgcolor'] = c1
c2 = random_color(h=0.4, l=0.7)
t.children[1].img_style['bgcolor'] = c2
# fix a reasonable size for the image, so tree is scaled to it
t.render('test.png', w=1600, units='px', tree_style=ts)
0
Entering edit mode
10.4 years ago
Daniel
★
4.0k
Check out http://topiaryexplorer.sourceforge.net/, it's designed exactly by this and is made by part of the QIIME team.
0
Entering edit mode
10.4 years ago
Sishuo Wang
▴
230
Thanks for all of you. It seems that Dendroscope is a nice choice after searching for a long time.
Similar Posts
Loading Similar Posts
Traffic: 2500 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Pointers are in similar question: What Is A Good Phylogenetic Tree Display Program For Large Data Sets?