Excuse me:
By far, I have found snp and indel variants in my whole exome sequencing.
I want to know how to find structural variants in the data.Are there any modules in GATK or third parties tools?
Thx in advance, I will be much appreciated.
Excuse me:
By far, I have found snp and indel variants in my whole exome sequencing.
I want to know how to find structural variants in the data.Are there any modules in GATK or third parties tools?
Thx in advance, I will be much appreciated.
If you are interested in GATK, this 3 years old answer is what you are looking for. GATK has [Genome STRiP] (http://www.broadinstitute.org/software/genomestrip/genome-strip) tool.
You can also play around with these tools ( https://www.biostars.org/p/77753/ ) (I would start with delly & pindel).
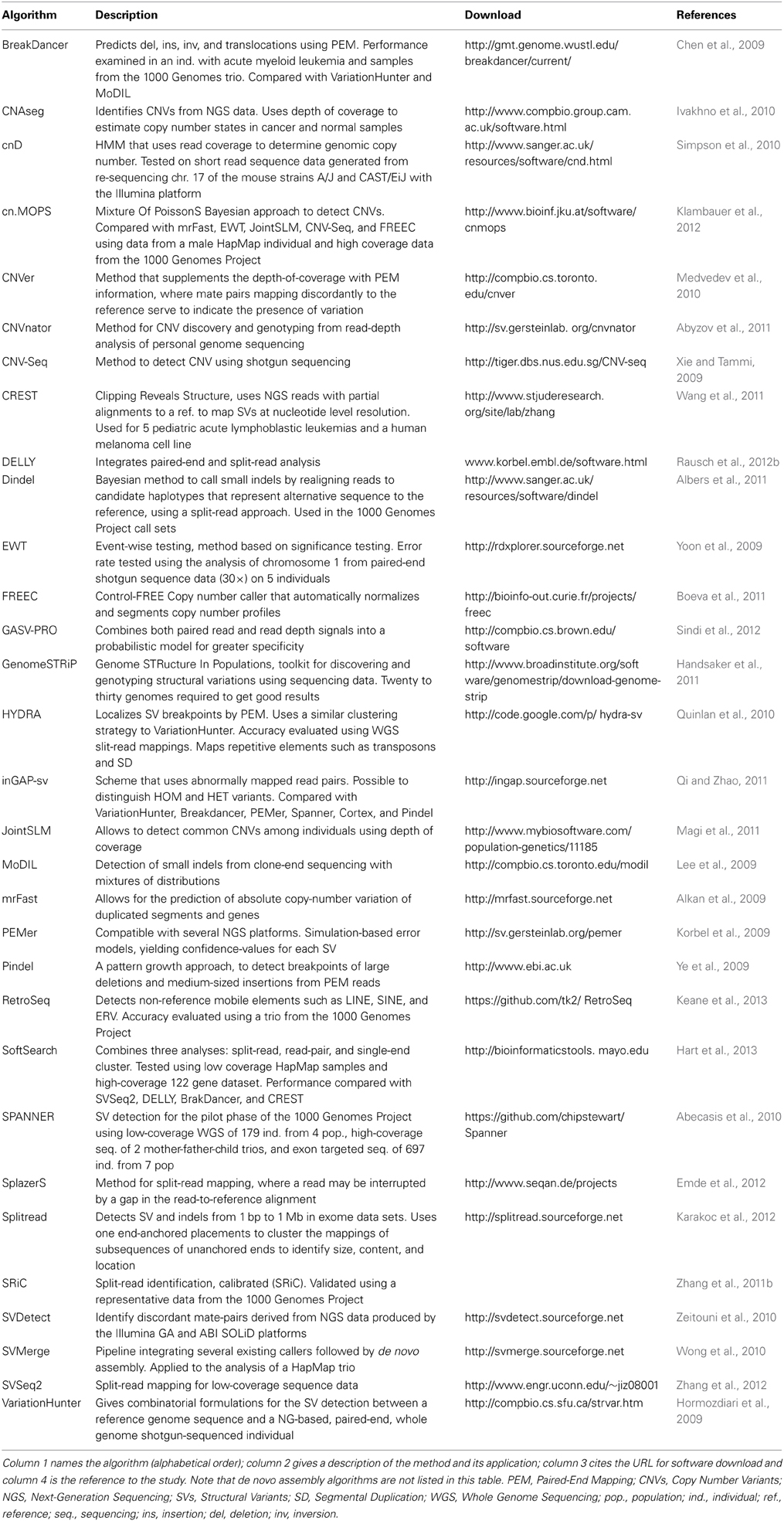
You can find even more tools here: Algorithms for the detection of structural variation. (Keane M. et al., "Identification of structural variation in mouse genomes", Frontiers in Genetics, 2014).
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.