A Beginner's Guide to NGS Data Analysis
Quality Control, Read Mapping, Visualization and Downstream Analyses



When?
9 - 13. March 2015
Where?
Leipzig, Germany
Scope and Topics
The purpose of this workshop is to get a deeper understanding in Next-Generation Sequencing (NGS) with a special focus on bioinformatics issues. Additionally, all workshop participants should be enabled to perform important tasks of NGS data analysis tasks themselves.
The first workshop module is an introduction to data analysis using Linux, assuring that all participants are able to follow the practical parts. The second module dicusses advantages and disadvantages of current sequencing technologies and their implications on data analysis. The most important NGS file formats (fastq, sam/bam, bigWig, etc.) are introduced and one proceeds with first hands-on analyses (QC, mapping, visualization). You will learn how to read and interprete QC plots, clip adapter sequences and/or trim bad quality read ends, get bioinformatics backgrounds about the read mapping and understand its problems (dynamic programming, alignment visualization, NGS mapping heuristics, etc.), perform your own mapping statistics and visualize your data in different ways (IGV, UCSC, etc.). The last two modules adress two specific applications of NGS: RNA-seq of model organisms and RNA-seq of non-model organisms.
Workshop Structure
The 2015 workshop has been redesigned and adapted to the needs of beginners in the field of NGS bioinformatics. The workshops comprises four course modules which can be combined.
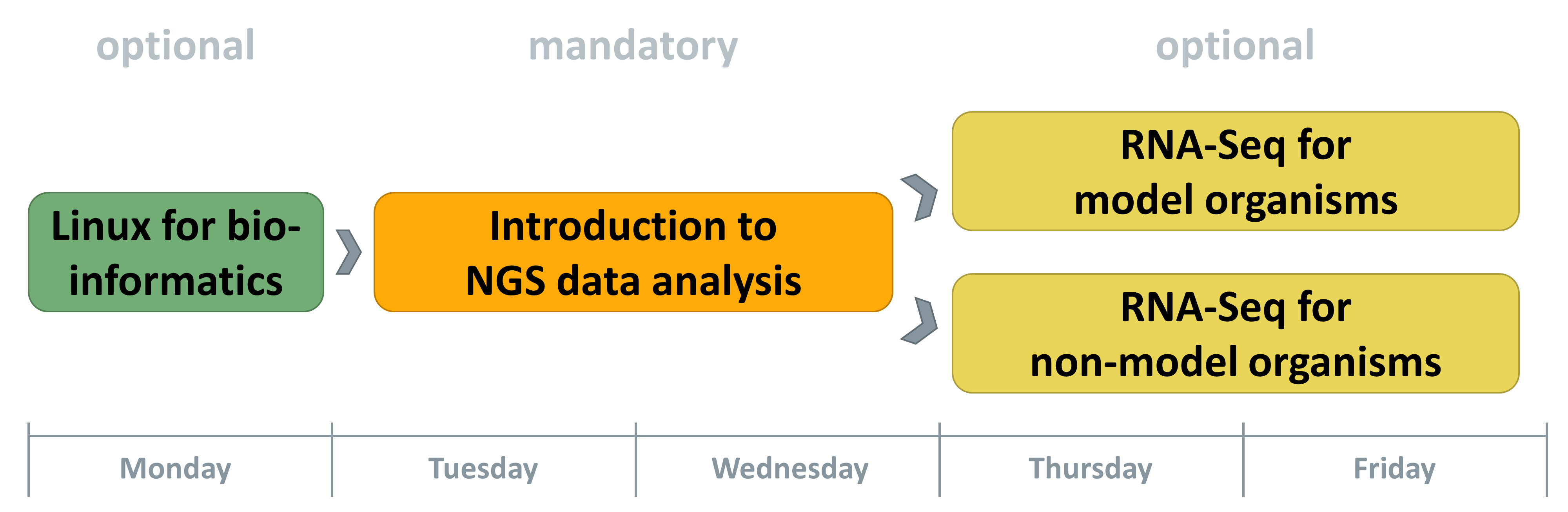
Linux for Bioinformatics:
This course module is optional. It will introduce the essential tools and file formats required for NGS data analysis. It helps to overcome the first hurdles when entering this (for NGS analysis) unavoidable operating system. Every participant who has no background in Linux usage should attend this course!
(The linux calls and commandline pipes teached here are the basis for all other courses and can not be covered again!)
Introduction to NGS data analysis:
This module is mandatory. Different methods of NGS will be explained, the most important notations be given and first analyses be performed. This course covers essential knowledge for analysing data of many different NGS applications. It also assures that all participants will be on the same level of knowledge for the downstream courses.
RNA-seq Data Analyses: Particpants can choose up to one of the following options:
- RNA-Seq for model-organisms
- RNA-Seq for non-model organisms
Depending on the organism you are working with, our trainers will show you what's possible with your data and how you could/should interprete the output data.
Key Dates
- Opening Date of Registration: 10 November 2014
- Closing Date of Registration: 1 March 2015
- Workshop: 9 - 13 March 2015 (8am - 5pm)
Attendance
- Location: iad Pc-Pool, Rosa-Luxemburg-Straße 23, 04103 Leipzig, Germany
- Language: English
- Available seats: 24 (first-come, first-served)
Course Prices and Program
The prices and the program can be found on our workshop website: http://www.ecseq.com/workshops/workshop_2015-01.html
Contact
ecSeq Bioinformatics
Brandvorwerkstr.43
D-04275 Leipzig
Germany
Web: http://www.ecseq.com
Email: events@ecSeq.com





Some updates: