Hi !
I'm currently working with paired-end RNA-Seq data from an oriented library (fr-stranded firststrand) of Salmonella enterica. Within this context, I'm performing some analysis with a tool developed in my lab that uses XS-strand attribute that TopHat adds in BAM files. It is known that Tophat is not designed to work on bacteria and returns poorer mapping results than Bowtie2 or STAR on those organisms (so far, my observations confirm this fact : ~65% uniquely mapped read with TopHat, ~80% to ~84% with Bowtie and STAR. Each time, the intron length was set to 0 or 1; because I suppose there is no intron in Salmonella, it's a bacteria).
My goal is to reproduce TopHat's XS-Strand attribute with a post treatment on a Bowtie/STAR mapping. I thought TopHat was using the SAM-Flag present in the second column of SAM/BAM files to guess the XS-Strand. To be sure, I ran the following command line:
grep -Po '\t[0-9]+\t.*XS:A:[\+\-]' accepted_hits.sam | awk '{print $1"\t"$NF}' | sort -n | uniq -c
Where accepted_hits.sam is the result of my TopHat run.
The following lines are an extract of the proportion of SAM-flags the previous command line returned:
NB Reads | SAM-Flag | XS-Strand attribute
262102 81 XS:A:+
59 81 XS:A:-
11563221 99 XS:A:-
1150 99 XS:A:+
11562241 147 XS:A:-
2130 147 XS:A:+
251 163 XS:A:-
9854904 163 XS:A:+
A read with a 99 SAM-Flag has the following characteristics (according to [1]): read paired, read mapped in proper pair, mate reverse strand, first in pair. I thought this SAM-Flag would refer to a + Stranded read ... But TopHat does not agree (11,563,221 times!). Notwithstanding this remark, a read with a 147 SAM-Flag has the following characteristics (according to [1]): read paired, read mapped in proper pair, read reverse strand, second in pair. I totally agree with the XS-Strand attribute predominant group, the read is on the - strand.
To be sure, I opened my BAM files with two genome viewers (Artemis and IGV) and had a look at my data. Both Genome viewers displayed the read orientation I was waiting for, and not the one TopHat wrote in its' 99s-XS-Strand attributes.
Here is a summary of my questions:
- With TopHat, one SAM flag can lead to both XS-Strand attributes (with a large predominance of one on the other). How is that possible?
- The predominant TopHat's XS-Strand-attribute group and the information given by the Broad Institute seems (sometime) contradictory. Which one should I trust?
If you have any questions, please feel free to ask. Many thanks in advance for your help !
PS : Here are the first lines of the accepted_hits.bam (obtained with: samtools view -h accepted_hits.bam | head). There is also my TopHat command line.
@HD VN:1.0 SO:coordinate
@SQ SN:gi|378697983|ref|NC_016810.1| LN:4878012
@PG ID:TopHat VN:2.0.9 CL:/usr/bin/tophat -p 5 -o ./Output_Salmonella/ --library-type fr-firststrand Index/salmonella.fasta_ref fastq/Salmonella_R1_egal.fastq fastq/Salmonella_R2_egal.fastq --min-intron-length 0 --max-intron-length 1
NS500446:4:H18FBBGXX:1:23206:24124:12692 153 gi|378697983|ref|NC_016810.1| 3 50 150M * 0 0 AGATTACGTCAGGTTGCAAGAGATCATGACAGGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCTCATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACA FFFF7FFFFF.7FFAFFF<FFF<F.F.FF7FFFFFFF.AFAFFF7FF<F77F<FF.<<F7F7FFAFFF)FF<FF7<FFFFFF)FF7FFFFFFFAFFFF.F)FFFAA.FAFFFAFAFFFAFFFFFFFFFFFF7F)F<FFFFAFFFFAAAAA AS:i:-5 XN:i:0 XM:i:2 XO:i:0 XG:i:0 NM:i:2 MD:Z:10T57A81 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:2:21104:23648:17357 153 gi|378697983|ref|NC_016810.1| 3 50 150M * 0 0 AGATTACGTCAGGTTGCAAGAGATCATGACAGGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACA FF<..AA)AF7FF)7FFFAFFFFFFFFFFF<F7<<<FFF<FFFF.AFAF<FAFA<.FAF7FFFAFFFFFFF.FFFF7FFFFFAFFFF<.F.FFFFFAA.FFAFFF7)FFAFFFFFFFFF<FFFFFFFFFFF7FFFF7F.F<FF.FAAAAA AS:i:-4 XN:i:0 XM:i:1 XO:i:0 XG:i:0 NM:i:1 MD:Z:10T139 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:1:12106:15808:8809 99 gi|378697983|ref|NC_016810.1| 4 50 30M1D120M = 39 185 GATTACGTCTGGTTGCAAGAGATCATGACAGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACAAG <AAAAFFFFFFFFFFFFAFFFFFFFFFFFAFFFFFFFFFFFFFAFFFFAFFFAFFFFFFFFF7FFFFF<FFFFFFFFFFFFFFFAAFFFFFFFFFFF<FFAFFFFAFAFFFFFFF<FAFFF.FFFFFFFFAFF<<<A.F.FFAFF<F77F AS:i:-8 XN:i:0 XM:i:0 XO:i:1 XG:i:1 NM:i:1 MD:Z:30^G120 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:1:12303:25690:20081 73 gi|378697983|ref|NC_016810.1| 4 50 150M * 0 0 GATTACGTCTGGTTGCAAGAGATCATGACAGGGGGATTTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAATTTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACAA <<AAAFFF7.FAFFFFAFFFF7FFFFFA.AFAFFFF.F.FFFF.FF<.A.<FAFAF)FFF)A7FFFFFFAFFFFA<FF.F<A7FFFFFFAAFF..AFF<.A.<AFFFAF7FFF<.F.A)<.F7.<FF)FA<F.F77FFF7<.FA)F<7.< AS:i:-7 XN:i:0 XM:i:2 XO:i:0 XG:i:0 NM:i:2 MD:Z:36A65A47 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:1:21202:15191:18222 99 gi|378697983|ref|NC_016810.1| 4 50 150M = 58 204 GATTACGTCTGGTTGCAAGAGATCATGACAGGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACAA AAAAAFFFFFFFFFFFFFFFFFFFFFFAFAFFFFFAAFFFFFFFFFAFFFFFFAFFF<FFFAFFAFFFFFFFFFFF<FFFAFFFAFFFFFFFF.AFFA7FFFFFFFAFFFFFFFAFFAFAFF7AFFFFFFFFFFF.<F)F7FFA.FAF7F AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:150 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:1:21302:7977:17040 99 gi|378697983|ref|NC_016810.1| 4 50 150M = 75 221 GATTACGTCTGGTTGCAAGAGATCATGACAGGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACAA <AAAAFFFFFFFFFFFFFFFFFFFFFFFFFFFFFFAFFFFFFFFFAFFF.FFFFFFFFAFFFFFFFFFFFFFFFF<AFFFFFFAFFFFFFFFF<FFFF.F<FAFF<7FFFFFF7FFFFA<FF7FFFFAFFFF<<F.AF7F.FF<AFFF.A AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:150 YT:Z:UU NH:i:1 XS:A:-
NS500446:4:H18FBBGXX:1:23312:4327:17892 99 gi|378697983|ref|NC_016810.1| 4 50 150M = 75 221 GATTACGTCTGGTTGCAAGAGATCATGACAGGGGGAATTGGTTGAAAATAAATATATCGCCAGCAGCACATGAACAAGTTTCGGAATGTGATCAATTTAAAAATTTATTGACTTAGGCGGGCAGATACTTTAACCAATATAGGAATACAA AAAAAFFFFFFFFFF.AFFAFAF<)FFA<AFAFFFF7FFFFFFFFFAFFAFFFFAFFFF7F)FFAFFAFFFFFFAAFFFF<FFFFFFFFFFF<.F.FF.FFFFF<AFA.FFF<FFFFF.F.F.F<FFFF.FFAFF..F7F.<FFFAF<FA AS:i:0 XN:i:0 XM:i:0 XO:i:0 XG:i:0 NM:i:0 MD:Z:150 YT:Z:UU NH:i:1 XS:A:-
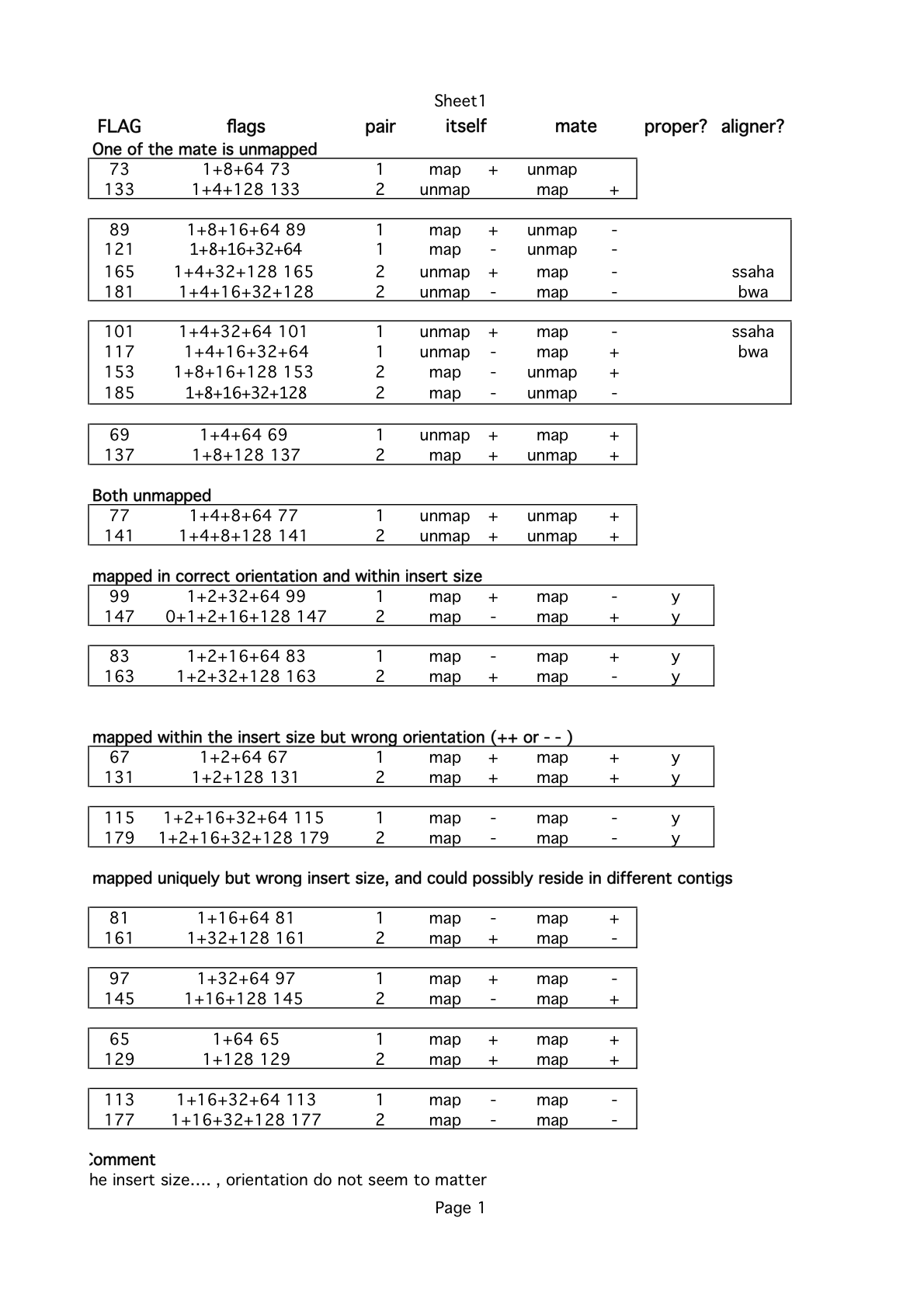
Here is a document about SAM-Flag interpretation I have found online. I mostly agree with the information given in this image: