Hi,
I have uploaded a code of diffpdb https://github.com/m4rx9/diffpdb.
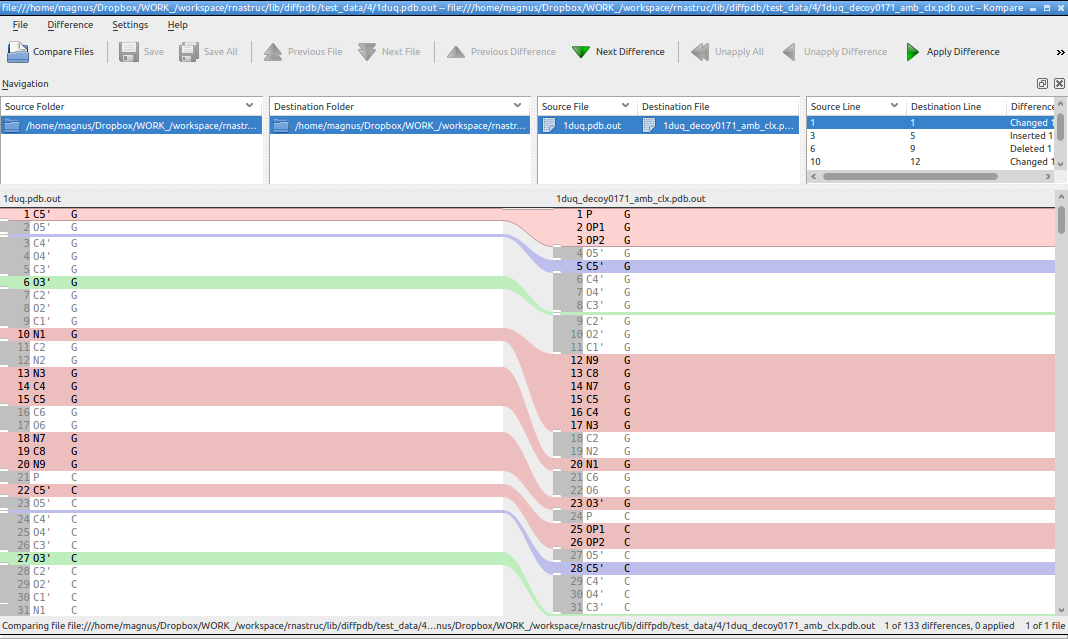
Working with pdb files can be a real pain. One problem is what are the difference between text content of two files. See the case below. For these two files is quite tricky to calculate RMSD and if you calculate RMSD line to line (atom to atom) you are just wrong.
How to get the sense what are the differences?
We do not have to reinvent the wheel :-)
I'm using diff or kompare (http://www.caffeinated.me.uk/kompare/) to compare two text-contents using only interesting columns (ignoring for example atom positions, they almost always will differ between files).
I'm using the code for months and it seems to be very useful for me. It might be useful for you so that's why I'm sharing the code.
Join the project, comment, clone, pull, enjoy!
./diffpdb.py --names test_data/4/1duq.pdb test_data/4/1duq_decoy0171_amb_clx.pdb
Magnus
$ ./diffpdb.py -h
usage: diffpdb.py [-h] [--names] f1 f2
positional arguments:
f1 file
f2 file
optional arguments:
-h, --help show this help message and exit
--names take only atom residues names

