Hi,
I've used the UCSC Table browser to extract the 3'UTR, 5'UTR, intronic, exonic and promoter coordinates (1000 bases upstream of TSS) for a list of candidate genes. I've noticed however that my exonic coordinates also reports the part of the UTR's position as part of the exon's coordinates.
e.g.
3'UTR exon
chr 1 215793960 215795149 + chr 1 215793398 215795149 +
I know they are attached when you view them in the Genome browser, but I need the the specific coordinates of just the UTR portion and I need the exon's coordinates that doesn't overlap with that of the UTR. Does anyone know how I can separate the two.
In some cases, it's even exactly the same.
e.g.
3'UTR exon
chr 1 93307322 93307481 + chr 1 93307322 93307481 +
Is there a plausible explanation for this? I know there are many different splice variants for the same gene, but it just seems really odd. I have 175 candidate genes and I see this occurrence quite frequently for the 5'UTR and its exon as well.
e.g.
5'UTR exon
chr 1 215747128 215747182 + chr 1 215787128 215787182
Thank you in advance
Tracey

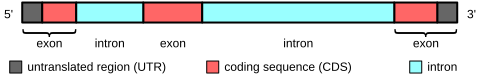

Since UTRs are expressed, they're part of exons, by definition.