Hi,
I have RNA sequencing data of T-Cell Receptor from several patients before and after immunotherapy. The sequencing was done by a vendor and they provided us with tables of data for individual patients. These tables contain rows with clonotypes (as per my understanding defined as CDR3 sequences with a unique combination of V, D, and J segments, along with counts of the clonotype. The CDR3 AA sequences are given in each row along with the V, D, and J names. The D segments are missing (I believe that should be fine). But I found some of the AA sequences are duplicates and so are some of the V-J segment combinations. This is confusing to me because each row should contain a unique clonotype. I am new to this type of data and might not understand this type of data correctly. Can someone please help me out with understanding this data set.
Thank you,
- Pankaj

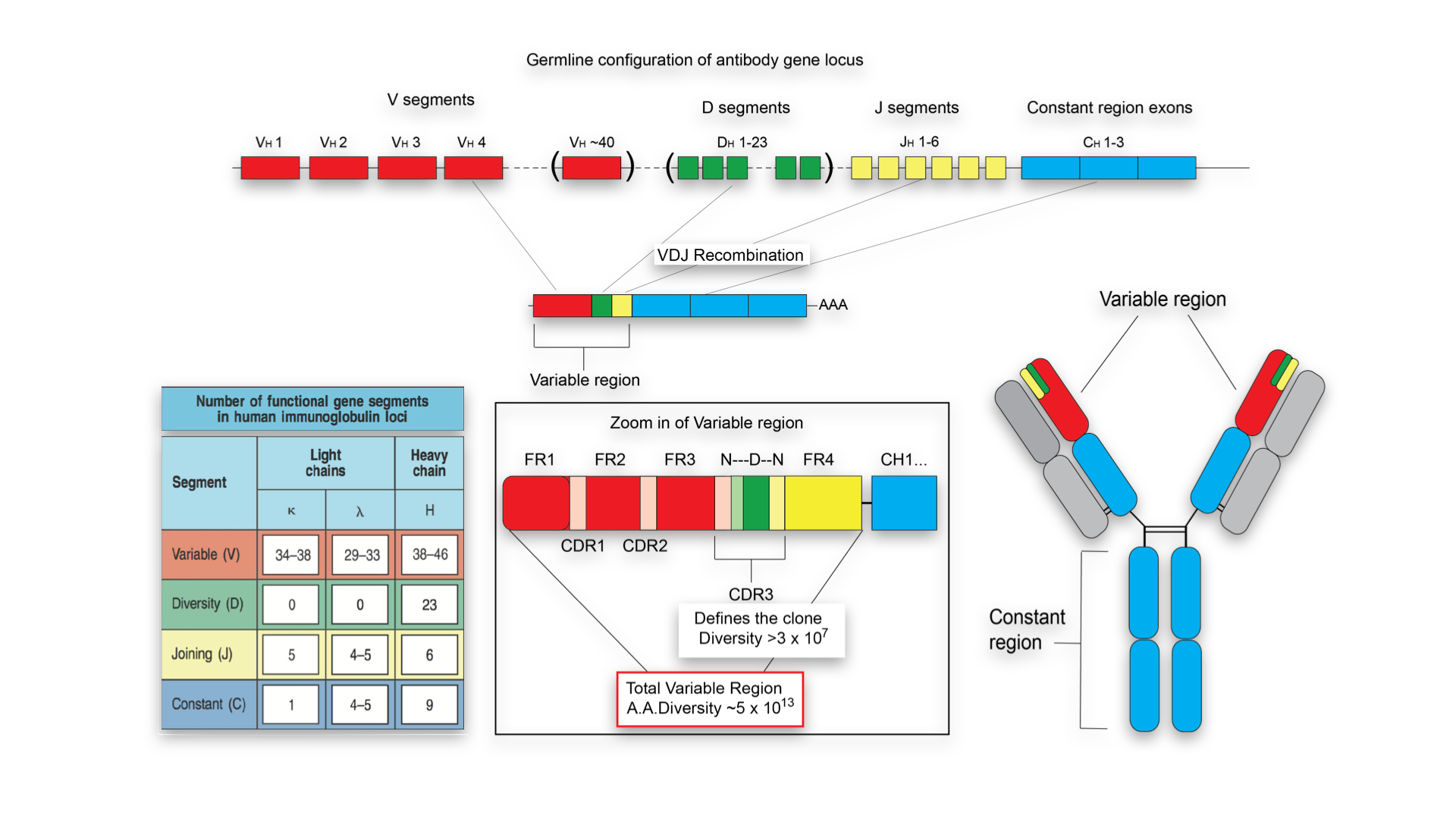

This was very helpful. I had a couple of follow ups: 1. Is there a reference for the figure and the second bullet point. I would like to use them for as a references in a paper we are working on. 2. How variable is the N region in terms of number of nucleotides.
Hello!
1a I've just referenced this page: https://www.bsse.ethz.ch/lsi/research/systems-immunology.html . I bet such sort of picture can be found in various textbooks, e.g. Janeway's immunobiology
1b http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2984183/ - for convergent recombination. In http://www.jimmunol.org/content/196/12/5005.short we have a figure directly showing that for the core (naive) repertoire the #nt sequences per #aa sequences is > 1.
2 Again you can check out the "insert size" figure from http://www.jimmunol.org/content/196/12/5005.short that was generated for healthy donors in various age. For a more detailed insert size distribution of V-D-J recombination machinery please refer to http://www.phys.ens.fr/~awalczak/PUBLI/mmwc12.pdf