With what software tool can I reconcile the gene tree (obtained for orthologs using OrthoFinder) versus the species tree for ~ 30 plant species?
I was thinking of using NOTUNG, but realized it may not output the specific image I am looking to generate.
Which is: ready-made tree that has the gain and loss numbers of the orthologs mapped on to internal nodes and terminal leaf nodes of the species tree.
Something like what I find in these example images :  , or
, or 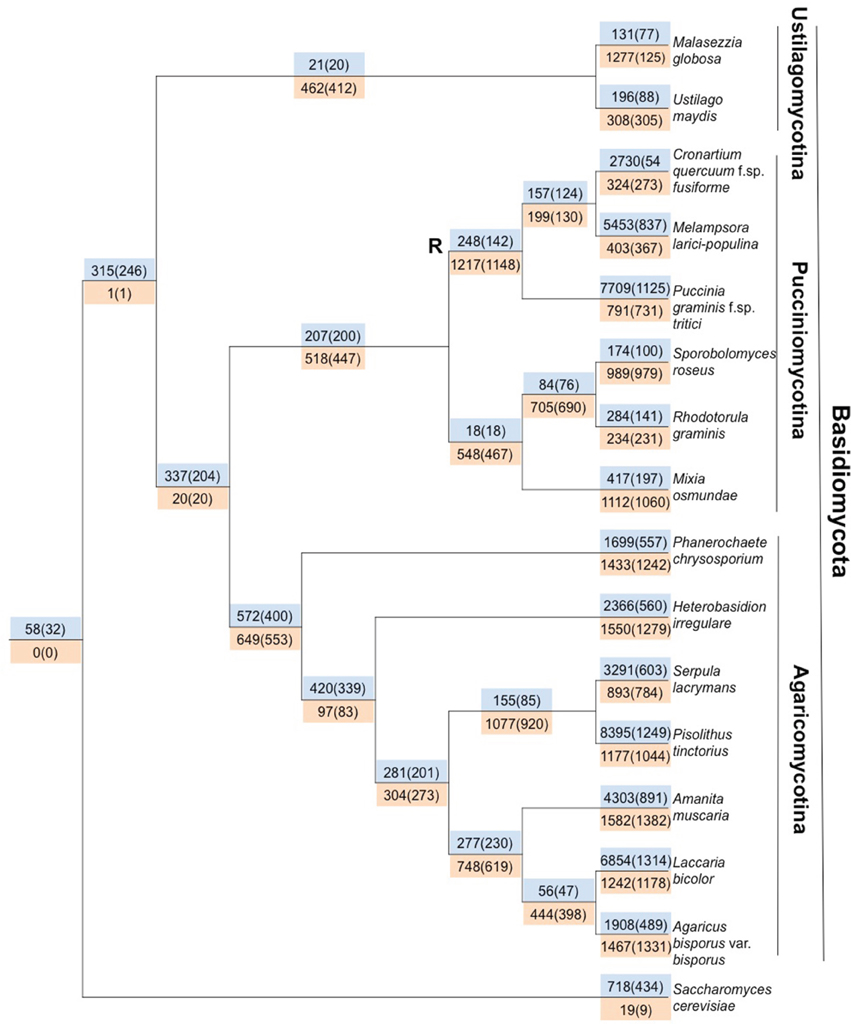 .
.
Please note that I am currently not using an outgroup species, or outgroup sequences for gene tree.


Did You ever find a solution to this ?
http://www.cs.cmu.edu/~durand/Notung/
Could you provide more information on how you ran Notung?