Hi there, we have now got the 12 sequencing samples back. However, when reading the FastQC report, we saw something weird: A large kmer content spike in the middle of the read.
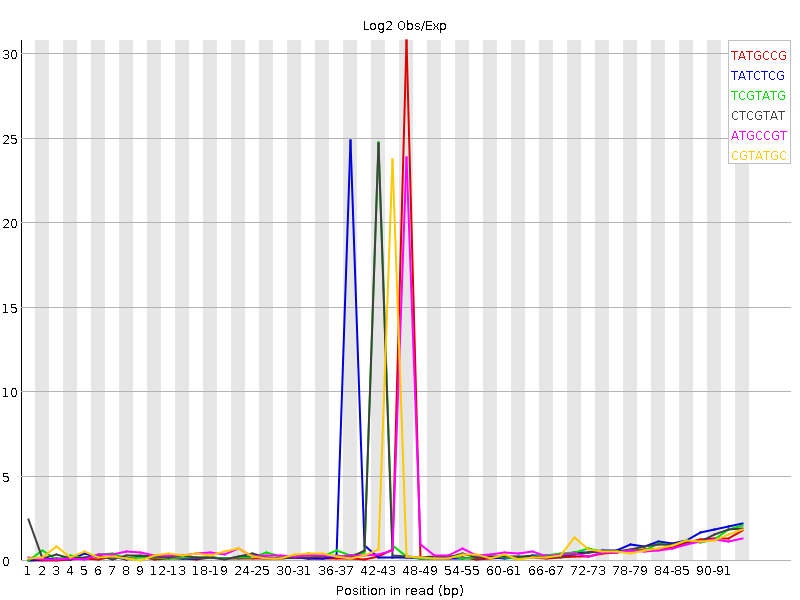
There is slight amount of adapter leftover but they should inflate the kmer content at the start of the read. Has anyone seen something like that before? Is it something to be worried about?
The full report for a pair of the reads can be found here. Thank you very much!


Unfortunately, that's like 3 years ago and I have no memory of that. I think @Ian's answer should most likely be what's happening
Ok, thanks anyway... so I guess, according to Ian's response, that's useless try to do some kind of trimming, isn't it? Thanks again!
Maybe just check the number of reads that show this kind of problem. If the number is small, just remove them