Hi i am new to R as well as microarray data analysis. I processed illumina array data using genomestudio and got the B-allele frequency and Log R ratio. Now i am trying to plot these values for all the chromosomes in a single plot using R. But i am unable to do it. Please if you guys have some suggestion then let me know. Thanks.
2
Entering edit mode
8.6 years ago
Michael
55k
See Drawing Chromosome Ideograms With Data one of the methods listed there should work for you.
2
Entering edit mode
7.0 years ago
bernatgel
★
3.4k
To plot data along the genome you can use karyoploteR. It's in Bioconductor (so you can install it with BiocManager::install(karyoploteR) and can plot almost anything on almost any genome.
There's a tutorial and examples page that contains a specific example on how to plot BAF and LRR values from SNP array that produces a plot like this:
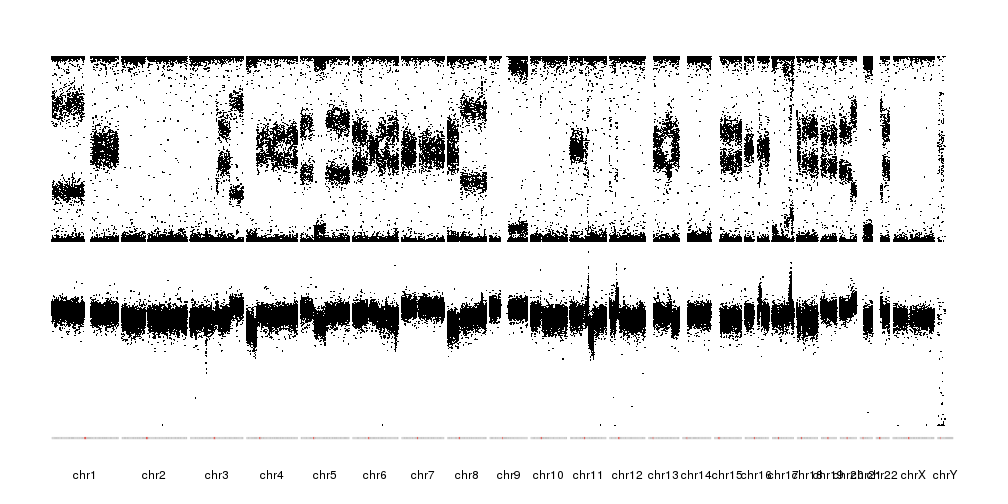
You can customize the image in many ways to adjust it to your needs.
Similar Posts
Loading Similar Posts
Traffic: 1509 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Thanks for the information