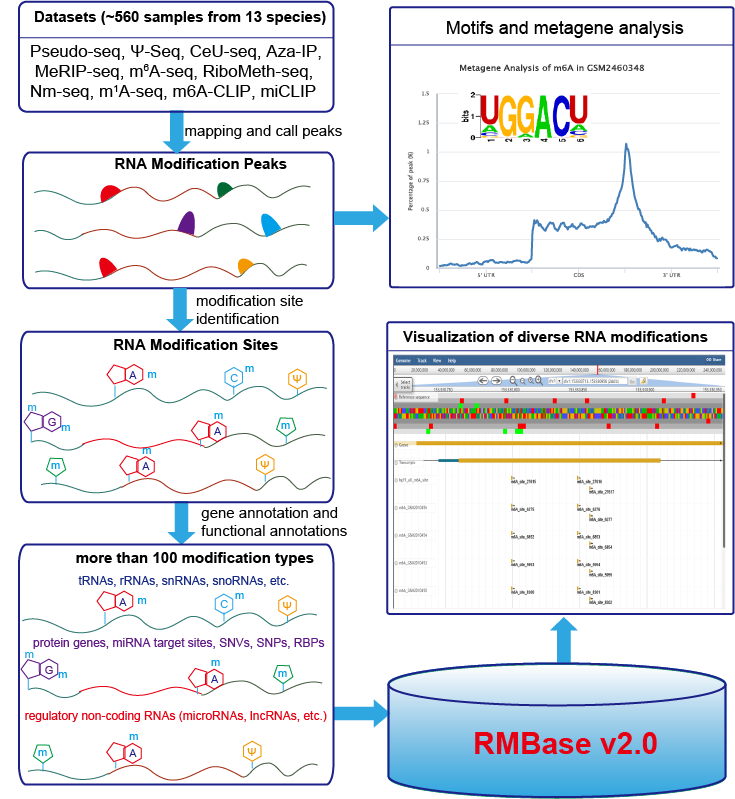
RMBase (RNA Modification Base) is a RNA modification database and designed for decoding the landscape of RNA modifications (>100 different modification types) identified from thousands of high-throughput RNA sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, miCLIP, m6A-CLIP, RiboMeth-seq) and public modification databases.
It contains ~226000 N6-Methyladenosines (m6A), ~9500 pseudouridine (Ψ) modifications, ~1000 5-methylcytosine (m5C) modifications, ~1210 2'-O-methylations (2'-O-Me) and ~3130 other types of RNA modifications(e.g. m1A, m6Am).
RMBase demonstrated thousands of RNA modifications located within regulatory RNAs (e.g. mRNAs, lncRNAs, miRNAs, pseudogenes, circRNAs, snoRNAs etc.), miRNA target sites and disease-related SNPs.
By integrating GWAS data into database, RMBase can be used to illustrate the clinically relevant RNA modification sites.
RMBase demonstrated thousands of RNA modifications located within miRNA target sites (~20,000) and RNA-Binding Proteins (RBPs) regions(>1,000,000 items).
This resource and tools will greatly help biologists to explore the prevalence, mechanism and function of various RNA modifications(>100 types) and link them with human cancers and other diseases.
RMBase has been updated to RMBase v2.0. RMBase Database is freely available at http://rna.sysu.edu.cn/rmbase/index.php.
RMBase paper is available at http://nar.oxfordjournals.org/content/44/D1/D259 and pubmed
RMBase continually be maintained and updated whenever novel high-throughput epitranscriptome sequencing data sets are released in public databases.


Hi lsp03yjh, I think this looks like a great resource for the community, but is there a reason you keep editing this post? It's not clear if you are adding new information that is necessary, in which case it is not helpful to keep bumping the post to the front. Sorry if I am missing something.
Thanks for your comments. I only want to update the number of m6A modification sites to ~226000 N6-Methyladenosines (m6A) that we have imported our resource.
Hi lsp03yjh, You really did a great jobs in Bioinformatics.
RMBase, deepBase, starBase, all of them are useful tools.
I will study them .
Thank you very much .
Thanks for your interest in our tools.
We also developed StarScan, snoSeeker/snoSeekerNGS, tRF2Cancer, ChIPBase, as well as starBase, deepBase and RMBase.
I am very interested in RNA biology and RNA informatics. Currently, we are developing novel experimental methods to explorer novel ncRNAs and their functions or mechanisms.