i have found 30-50 bp is the length of short reads but do not know about long reads......... what is the appropriate length of long reads in an average???
See this nice graphic:
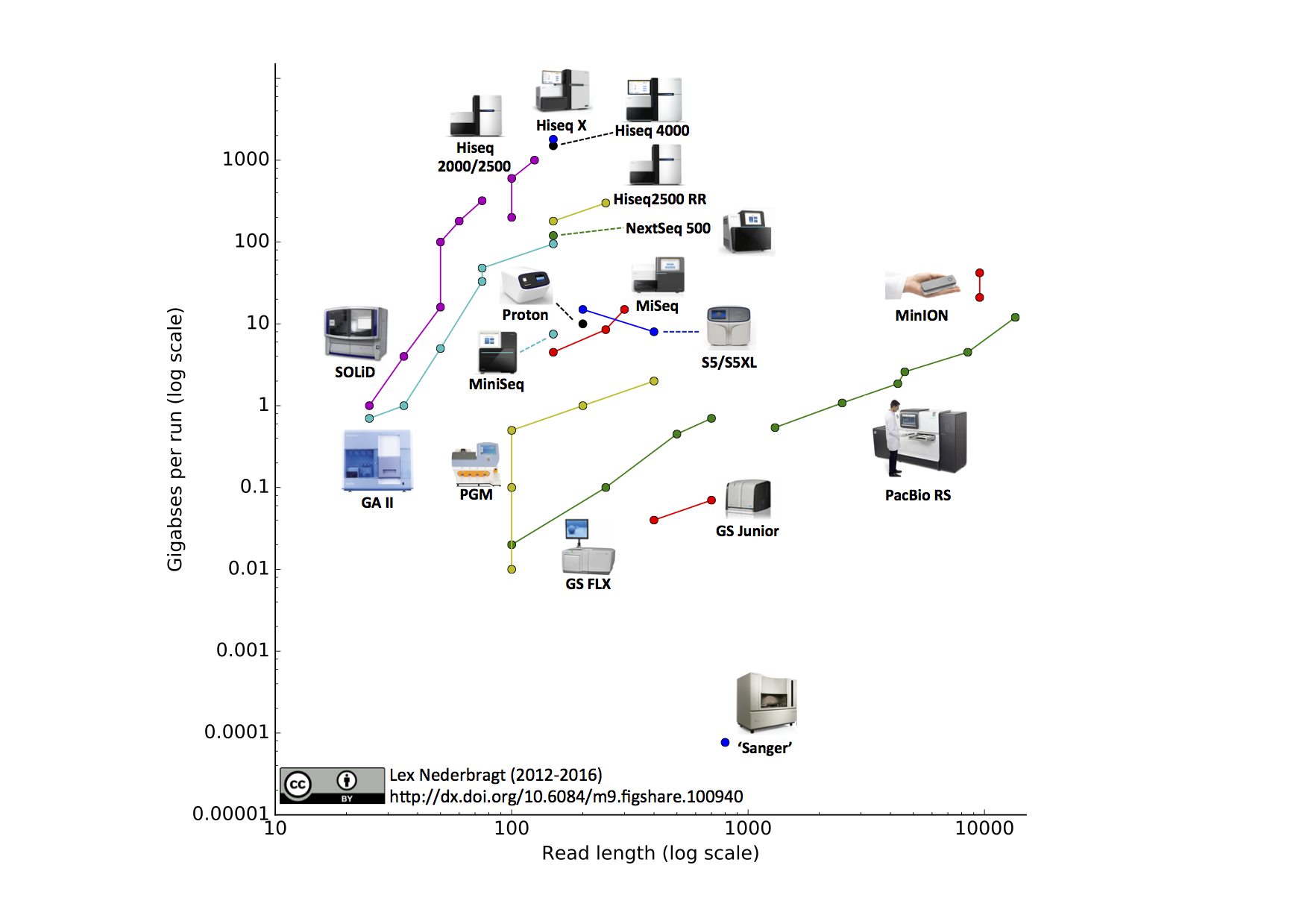
30-50 bp is very short, inappropriately short for most applications. Long reads on PacBio platform are ~12kb if I'm not mistaken, reads using Oxford Nanopore sequencers can be tens of kb's (easily 20kb) and maximum mapping reads that has been reported is >150kb.
Sure. Specific fields can vary. Some fields are also okay with Sanger. I meant overall.
You can check something like SRA for exact stats.
Or you can do a back of the envelope calculation. Most sequencing is Illumina. Most Illumina sequencing is HiSeq. Most HiSeqs do not support 150bp reads.
30-50 bp is very short, inappropriately short for most applications.
Not sure I agree with that... For ChIP-Seq and RNA-Seq for gene expression (no de novo assembly or splicing) the advantage of going above 50 I think is quite small. Also for bisulfite sequencing above 50 you don't gain much in mappability. (I'm referring to mouse or human genomes). The of course, the longer the better...
Of course other things being equal you would go for longer reads. Still I'm quite convinced that in most differential gene expression experiments 50 bp reads are going to be substantially similar to 75+ bp. The gain in better alignment is probably minimal. If for example money is a limiting factor, I would definitely prefer to sequence shorter reads but do more replicates or more meaningful follow up experiments. If tophat2 "refuses" to align shorter reads, which I kind of doubt, just choose a different aligner. Again, I'm just talking in general terms...
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


from which sequencing platform? and how did you found that 30 bp is length of short reads?
30-50 bp is where we started at a decade ago. We are way past that stage now even on the short read technologies (e.g. Illumina, Ion).
With technologies like 10x genomics/Illumina sequencing, the reads (contigs that come out of the process) can be on the order of hundreds of kb.