I would like to know how I can use SOM for disease detection. Given a lung cancer dataset, how can SOM be applied for detection, there are certain terminologies like, sensitivity, specificity and accuracy percentages....are there ways to calculate all these with the SOM algorithm? I would appreciate answers from anyone who can shed more light on this
See Self-Organizing Map (Wikipedia) for a brief description for SOM.
SOM like other members in the class of learning algorithms, requires a training and mapping (testing)data set. You can take 3/4 of your dataset for training and 1/3 for mapping. As you are now dealing with known datasets, you can use several measures like sensitivity, specificity and accuracy for the prediction assessment. Means, you can check how good is your prediction model using FP, FN, TP & TN from the results of your testing and mapping results. A PubMed search can get your several studies that applied SOM (or other learning algorithms) applied to problems in life science data with these parameters.
See Sensitivity and specificity (Wikipedia) for more detailed description on Sensitivity and Specificity
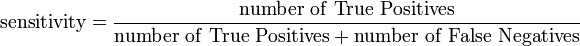
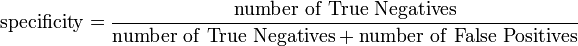

Khader gave an excellent answer on the use of SOMs for classification. I would like to complement his answer by pointing the reader to work done in the group where I did my PhD at the Radboud University in Nijmegen/NL, where a supervised self-organizing map was developed. The traditional Kohonen map is an unsupervised training method, though LVQ and other approaches have been used to make them more supervised. However, as far we we knew at the time, no true supervised learning of the class information was done then.
The R package kohonen implements this supervised SOMs as XY-Fusion maps. The training of the maps is very traditional, but the difference lies in the fact that the winning unit is determined by likeliness in the classification variable too (Y, or dependent variable). BTW, the method is not limited to one Y variable, and neither to a classification variable. I used various combinations in crystal structure classification problems in this paper:
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Hi Tim, sorry to say I do not want to put this into an answer because it is not really one. Most of the terms you are mentioning do not apply to self-organizing maps. Also youur question does not make much sense: you want to detect cancer in a cancer dataset ??? nothing to do! You should study the literature about classification, SOMs and study the nature of your data better. then you can come up with a better question.
I agree with Michael, But this is probably first question on Machine learning in BioStar. I am sharing some of my thoughts in the answer section. But you definitely get more help if you can provide us more details as suggested by Michael.
I agree with Michael. But this is probably first question on Machine learning in BioStar. I am sharing some of my thoughts in the answer section. You can definitely get more suggestions if you can provide us more details regarding your problem.
@Michael, why would you not be able to define measures for sensitivity, specificity or accuracy with SOM??
Because SOM and training algorithms as defined by Kohonen are unsupervised learning algorithms as you also write. Unsupervised learning does not include class-labels. That means that these measures do not apply directly. A SOM can be turned into a classifier or predictor by partitioning the output nodes. I would suspect that such a classifier and also the supervised SOM would be outperformed by a pure classifier (e.g. SVM) with real data. A comparision would be interesting.