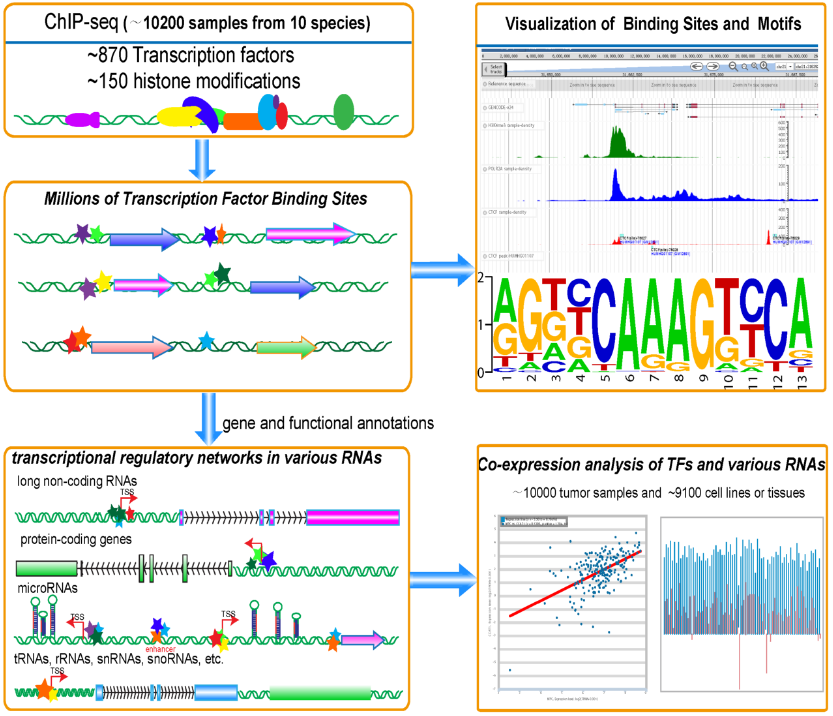
ChIPBase v2.0 is designed for studying transcription factor binding sites and motifs, and decoding transcriptional regulatory networks of non-coding RNAs (lncRNAs, miRNAs, snoRNAs, tRNAs...) and protein-coding genes (PCGs) from ~~10,200 ChIP-seq data (ChIP-seq, ChIP-exo, MNChIP-seq) in 10 speices (Human, Mouse, Fruitfly, Worm, A. Thaliana (plant), Yeast, Rat, Zebrafish, X. tropicalis, Chicken).
We identified thousands of Binding Motif Matrices and their binding sites from ChIP-seq data of transcription factors and predicted millions of transcriptional regulatory relationships between transcription factors and genes.
We constructed Regulator module to predict hundreds of transcription factors that were involved in or affected transcription of ncRNAs (lncRNAs, miRNAs, snoRNAs, tRNAs...) and protein-coding genes.
Moreover, we built a web-based tool, Co-Expression, to explore the co-expression patterns between transcription factors and various types of genes by integrating the gene expression profiles of ~10,000 tumor samples (TCGA) and ~9,100 normal tissues or cell lines.
ChIPBase also provides a ChIP-Function tool and a genome browser to predict functions of diverse genes and visualize various ChIP-seq data. This study will greatly expand our understanding of the transcriptional regulations of ncRNAs and protein-coding genes.
ChIPBase paper is available at http://nar.oxfordjournals.org/content/early/2016/10/20/nar.gkw965.full
ChIPBase is freely available at http://rna.sysu.edu.cn/chipbase/index.php


As far as I can see, the website is down.
Thanks your information. I test our website, it is ok, you can visit it again.
It still looks down to me, and I am not the only one: http://www.isup.me/rna.sysu.edu.cn - firewall issues perhaps?
Thank you. We are testing network, some regions can visit it, as you said that it might be firewall issues.
How does Chipbase compare to I-Cis-Target?
As far as I know, ChIPBase has much more available datasets than I-Cis-Target, and more over, ChIPBase provided a web-based tool named "Co-Expression" to explore and validate the co-expression patterns between transcription factors and various genes. And with ChIPBase, you can easily find TFs that bind at upstream and downstream of genes interested.
Thank you. Are you able to quantify "much more" available datasets? Also, for balance I should add that I-cis-target also has web-based tool that I'm about to use....hence the question how do they compare.
Hi ...I am interested in AR in breast cancer datasets. AR is not available in UCSC. Chip base is quite helpful.But, I do not know how can I generate image for this tables. This is giving me data for each dataset . How can i show multiple datasets into one figure.
http://rna.sysu.edu.cn/chipbase/regulation_locus_browse.php?organism=human&assembly=hg38&tf_symbol=AR&sample_id=HUMHG02225&stream=up-30kb&target_symbol=hsa-mir-17&ref_gene_id=MI0000071
Please open a new question for this, providing some background information and showing what you've tried so far. The more effort you show, the better the odds people are willing to invest time and effort into your question.