Does anybody know of numerical descriptors of nucleotide sequences, similar in idea to protein sequence descriptors (such as the sets described at http://www.biomedcentral.com/1471-2105/8/300). A trivial example would be a 4-element vector of the nucleotide frequencies.
There is a number of different DNA properties that can be estimated from the nucleotide sequence. Most of these are calculated from di- or tri-nucleotide lookup tables:
- Base-stacking energies (Ornstein et al. 1978)
- Nucleosome position (Satchwell et al., 1986)
- DNAse I sensitivity (Brukner et al., 1990)
- Intrinsic curvature (Shpigelman et al., 1993)
- Propeller twisting (el Hassan and Calladine, 1996)
- Deformability (Olson et al., 1998)
I was heavily involved in a few projects that made use of these parameters, in particular, for constructing visualizations of prokaryotic genomes (the GenomeAtlas method).
Regarding representation of DNA sequences, my experience is that you can get quite a lot more out of a nucleotide sequence by looking at di-nucleotides instead of individual nucleotides; a lot of the structural properties of DNA are primarily determines by which nucleotides are stacked on top of each other in the double helix. Taking into account also tri-nucleotides does not seem to add nearly as much in that respect.
One thing to keep in mind is that many of these are highly correlated with the AT content of the DNA sequence, and hence that the different DNA structural properties tend to also correlate with each other. Below are some scatter plots from an appendix of my M.Sc. thesis:
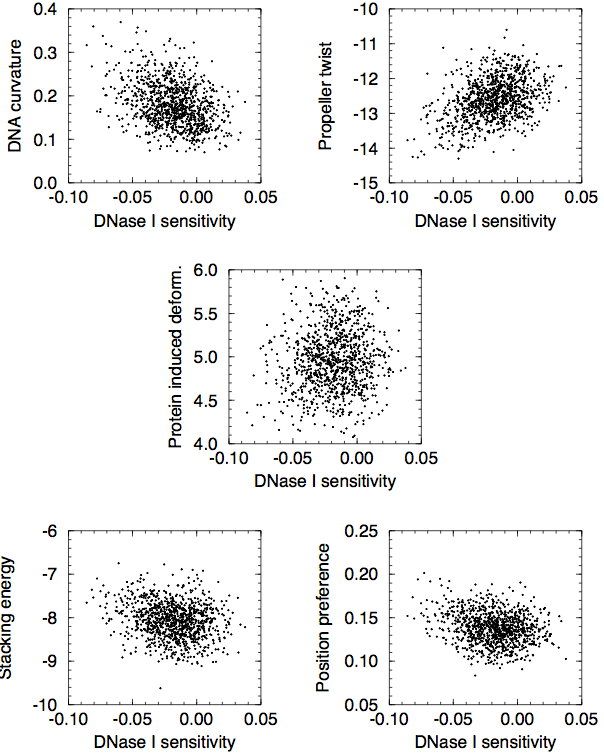
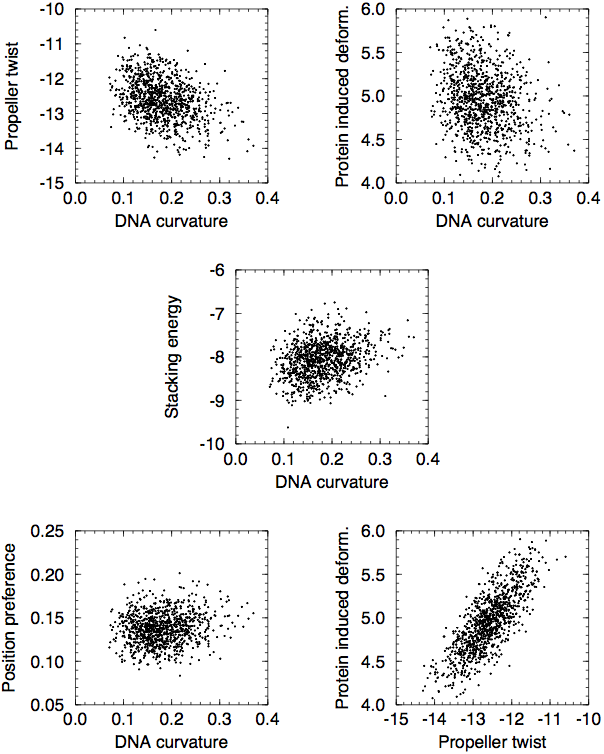
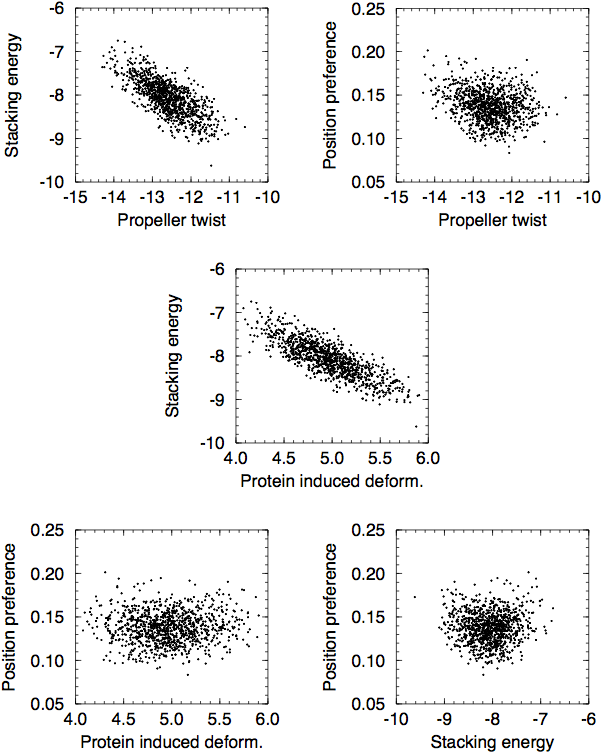
Finally, one should note that the nucleotide composition of protein-coding regions is very different from that of non-coding regions. This is one thing that can heavily bias any comparison of two sets of DNA, and it is thus important to make sure that any comparison is made between sets of DNA that contain the same fraction of protein-coding DNA.
I've seen numerical vectors used for nucleotides when, for example, developing SVM models for nucleotide sequence features. An example would be this study: "Prediction of polyadenylation signals in human DNA sequences using nucleotide frequencies". It's probably easiest to quote from the methods section:
Binary pattern: In the case of binary pattern each nucleotide was represented by a vector of four dimensions such as A by [1,0,0,0], C by [0,1,0,0], G by [0,0,1,0] and T by [0,0,0,1]. Thus a sequence of 200 nucleotides was represented by a vector of 800 (4 × 200) dimensions, which means UP100 and DW100 were both represented by vectors of 400 (4 × 100) dimensions.
Simple nucleotide frequency: In this case we calculated nucleotide frequencies of 100 upstream (UP100) and 100 downstream (DW100) positions, relative to poly(A) signals, separately and further added them to one another so that the total dimension is double. For instance, the sequence of 100 upstream was represented by a vector of four dimensions using mononucleotide frequency (frequency of A, T, G and C). In the case of dinucleotide frequency (AA, AC, AG, CG, AT ..), the sequence was represented by a 16-dimensional vector. Similarly, the sequence was represented by a vector of 64 dimensions in case of trinucleotides and by a vector of 256 dimensions in the case of tetranucleotides.
I think it's an approach used more commonly for protein sequences, because amino acids have more physicochemical properties that can be described using vectors.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Can you elaborate on what are you trying to use this data for? Also, I do not really see why the paper is trying to predict protein families with autocorrelation, when there are more promising (structure- and sequence homology based) methods available.
Decriptors = features derived from sequence or structure data ?
from sequence data