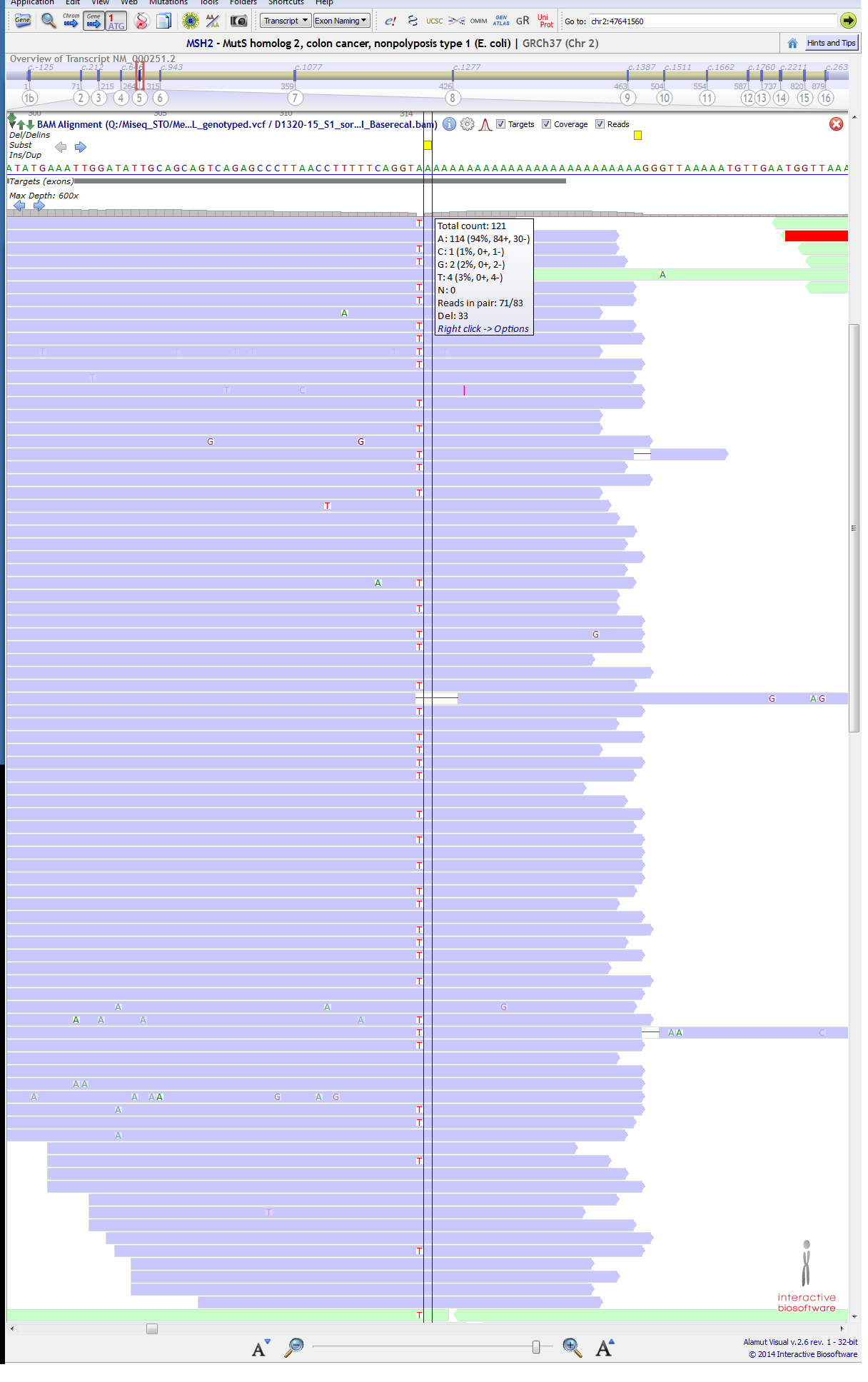
A variant is not getting called at position in MSH2 gene at ch2:47641560 (c.942+3A>T) It is a known pathogenic mutation. ( https://www.ncbi.nlm.nih.gov/clinvar/variation/36580/ ) and this variant have been mention in literature for not getting called ( http://www.sciencedirect.com/science/article/pii/S152515781630143X ), they say that "located in the 3′ end of exon 5 in a difficult-to-sequence homopolymer stretch of 27 adenines" so, I am OK with the fact that my VC pipeline is not calling this variant in a specific sample. (though T being 42% and most of them passing the phread-quality score). We have verified this variant in the same sample through Sanger sequencing. below are attached 1st & 2nd Image, Purple:forward strand, green:reverse strand).
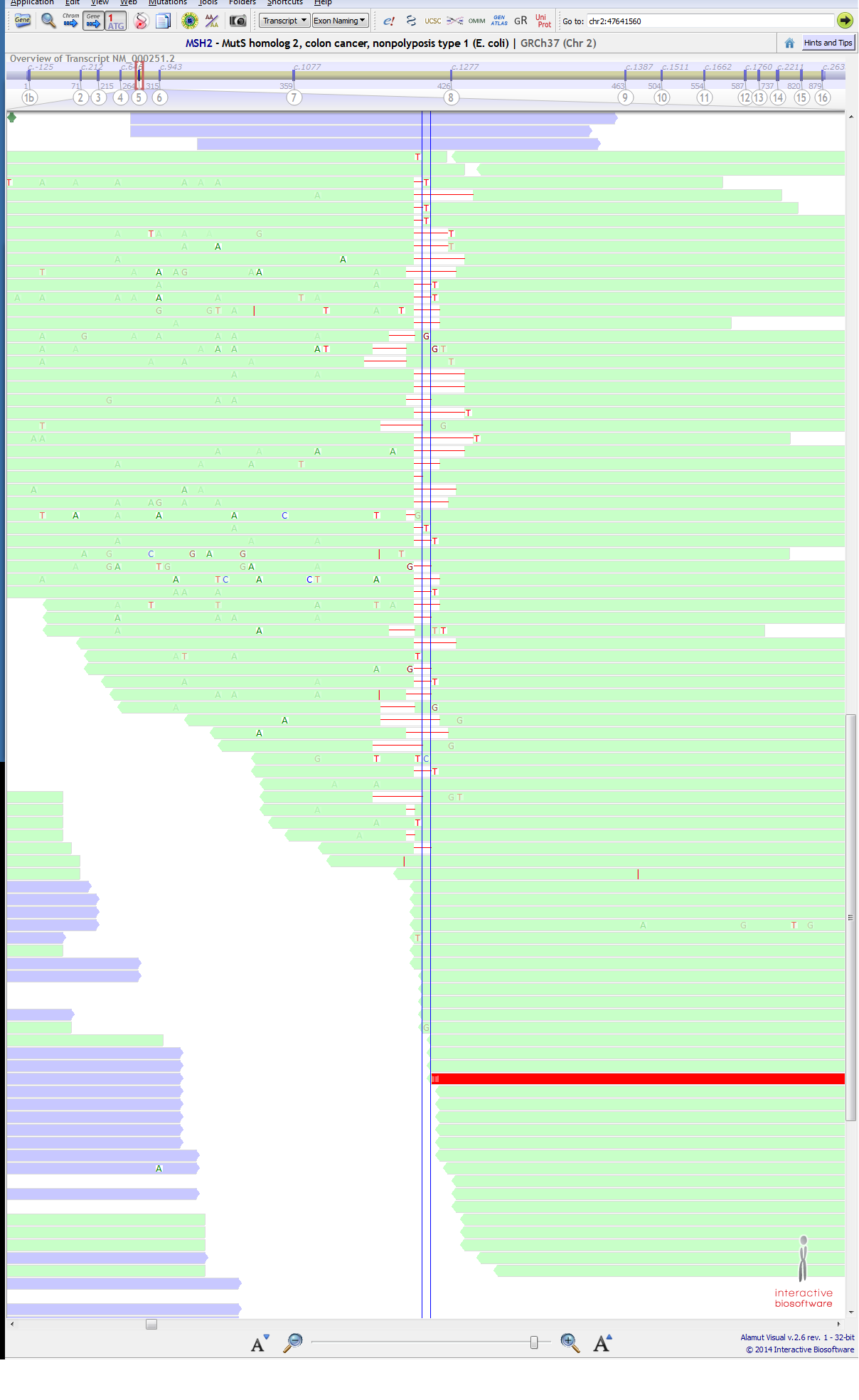
BUT....................... just next to this position at ch2:47641561 (c.942+4A>T), a variant is getting called for the same sample, though T is only 3% (4/121) at that position. below are attached 3rd & 4th Image, (purple:forward strand, green:reverse strand). I could not understand the reason... CAN ANYONE EXPLAIN IT??


Thanks & Happy Chrismas
Ashish


It looks like a single base heterozygous deletion from the first screenshot rather than an A>T change.
see 1st &2nd images (append) as one and 3rd and 4th as one (could not took sot of them in single frame) may be this story can help, http://gatkforums.broadinstitute.org/gatk/discussion/comment/35082
Thanks