I've noticed that most of the major projects involving wide-scale ChIP-seq analysis like ENCODE and Blueprint use a pipeline that involves running the SPP peak caller to estimate fragment size, then using Macs2 to do the peak calling but with the "--no-model" flag and giving Macs the fragment size estimated by SPP. Does anyone know the reasoning behind this? Is there a paper that shows the method used by SPP to estimate fragment size (cross-correlation) is more accurate than what Macs2 does? And how big of an impact does this have on the peak calling?
It is generally set to --nomodel for histone marks and ATAC, not for TF.
The macs2 requires a prior width to extend the reads to the fixed fragment size (--extsize) when --nomodel is set.
This will be set to a number that was estimated by phantompeaktools (run_SPP.r)
This will not have a very big effect for broadpeaks. The default parameter, I think it is 200 and it's should be fine in general.
Running SPP not only gives you the estimated fragment size, but also the NSC and RSC values, which tells the quality of chips. So people tend to run SPP to know the quality of chip and use the estimated fragment size while calling peaks using macs.
P.S: The run_SPP.r sometimes returns multiple estimated fragments lengths and the highest one or average is taken.
In my case, I can ask the lab that did the sequencing, and they can tell me EXACTLY what the fragment size distribution is. I would try talking to them. The fragment size reported by the lab that did the PCR is undoubtedly going to be better than any software estimate.
Advice for setting parameters: I almost tried (wherever possible) to use below mentioned parameters from ensembl for broad peaks using different peak callers. Check the results below (attached, all these peaks passed FDR < 0.05).
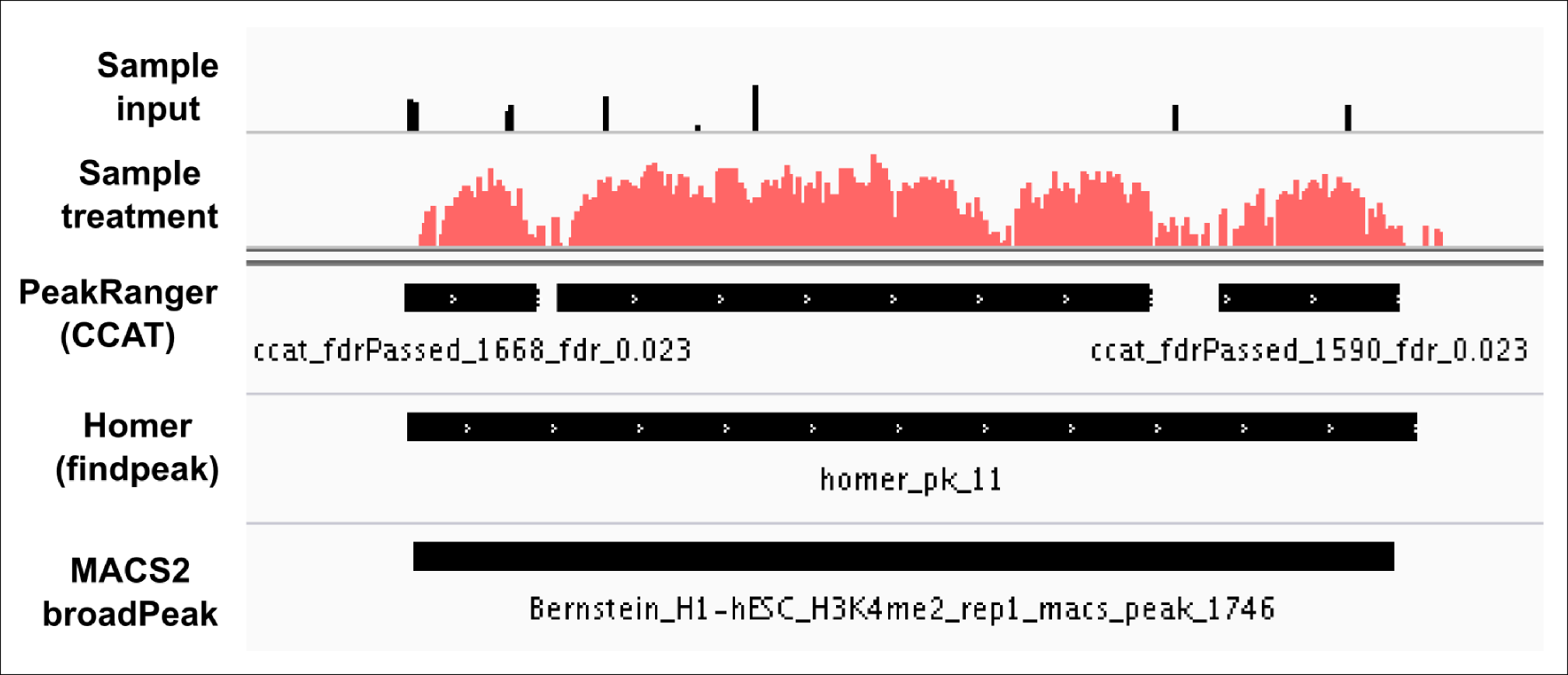
"The algorithm described by Xu et al, is specifically designed for peak calling of broad features, such as H3K36me3. The parameters were set empirically to (fragmentSize 200; slidingWinSize 150; movingStep 20; isStrandSensitiveMode 0; minCount 10; outputNum 100000; randomSeed 123456; minScore 4.0; bootstrapPass 50)."
SOURCE: http://www.ensembl.org/info/genome/funcgen/regulation_sources.html
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.

