Although it was released last year, deepTools2 has not been formally introduced to the biostars community yet.
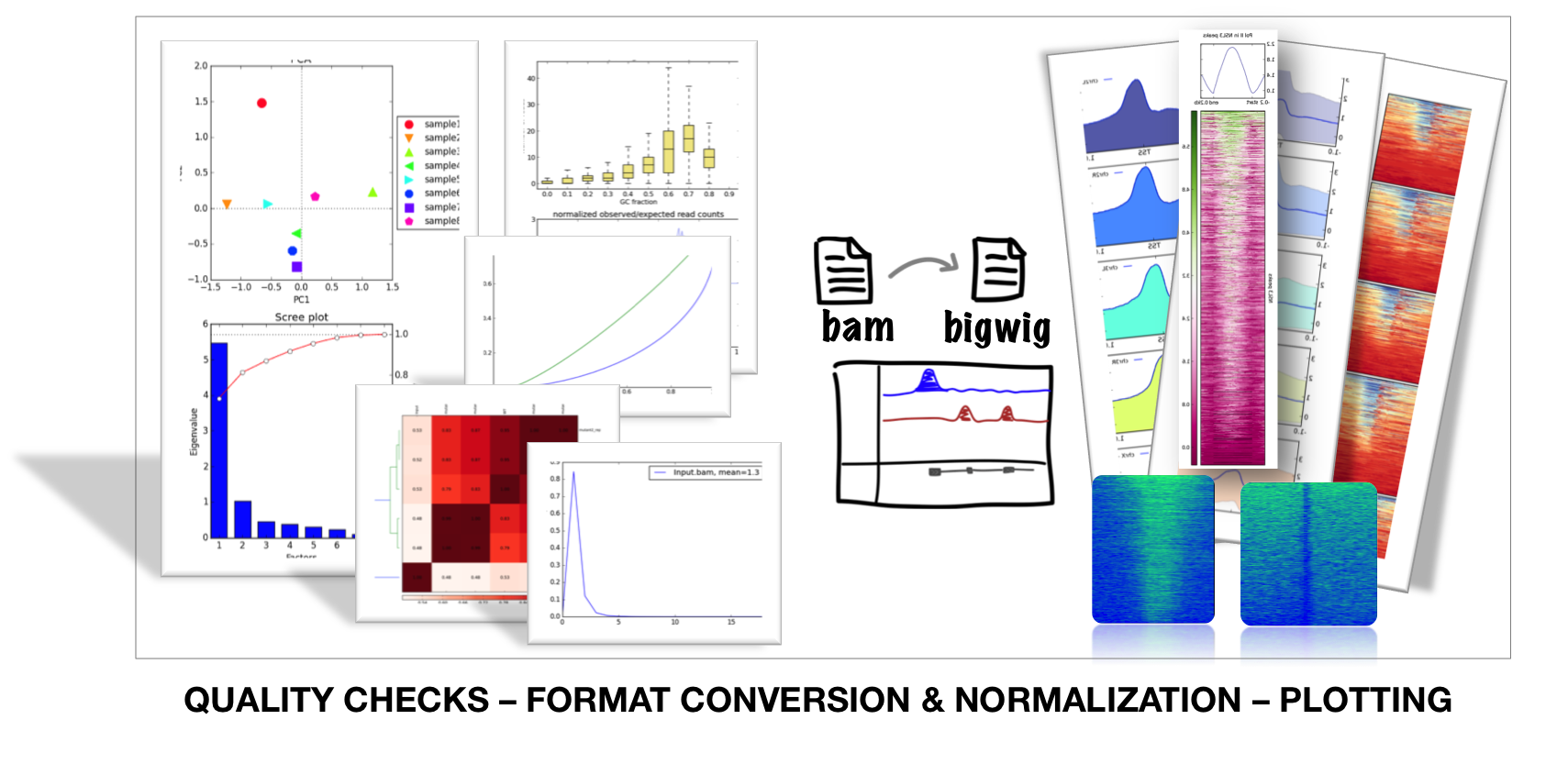
deepTools is a set of command line / galaxy tools for QC, normalization and visualization of sequencing data
deepTools addresses the challenge of handling the large amounts of data that are now routinely generated from DNA sequencing centers. deepTools contains useful modules to process the mapped reads data for multiple quality checks, creating normalized coverage files in standard bedGraph and bigWig file formats, that allow comparison between different files (for example, treatment and control). Finally, using such normalized and standardized files, deepTools can create many publication-ready visualizations to identify enrichments and for functional annotations of the genome.
Publication: https://doi.org/10.1093/nar/gkw257
Documentation: http://deeptools.readthedocs.io/en/latest/
Galaxy: http://galaxy.ie-freiburg.mpg.de Also a part of https://usegalaxy.org/
Code: https://github.com/fidelram/deepTools
Biostars: #deeptools
Lead authors are quite active on biostars (especially Devon Ryan).