========
Hi there, let's have a look at the structure of human genome first.
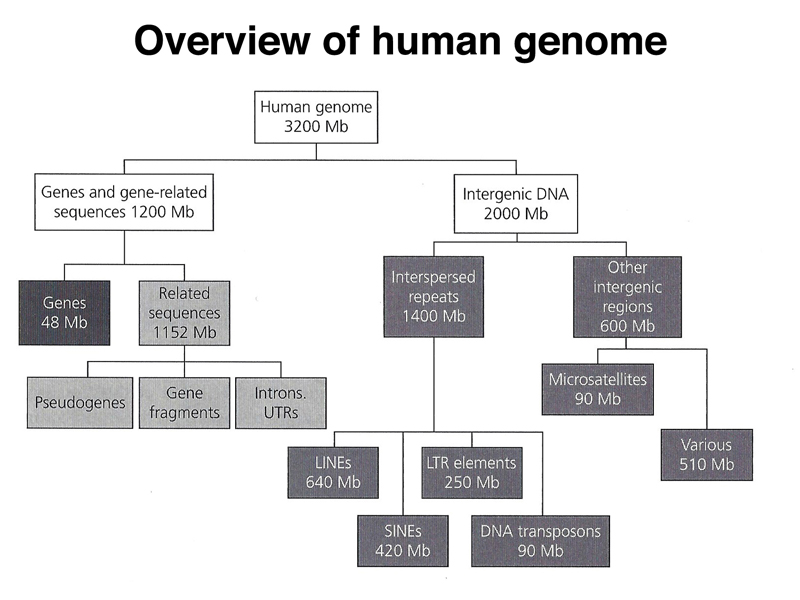
Now, given a sequence, referred to human reference genome hg19,
chromosome, start, end, seq
19, 41808719, 41808745, GGTGGTGGTGGCTTCCGGGGCCGCGGG
I want to know which component of the figure above this sequence belongs to. Could anyone tell me how to? Thanks in advance.
============== update ============
I just found ANNOVAR, it can do region-based annotation, I think it will work for me. I recommend it to others who may face the same problem as me.


Maybe blast2GO can help? I do not think it's a trivial task to map a sequence to its structure/function. Also, I'm assuming this is from human chr19.
You could always look for annotations by the chromosome coordinates.
Thanks @Ram , it's from chromosome 19 of hg19. I want to categorise sequences based on their feature. Now I am doing pilot study to find a suitable way to categorise them. Suggestions are welcomed.