Entering edit mode
7.6 years ago
Are there regions in the genome that are not covered by DNA sequencing?
If the genome would be a landscape, and you would create a map of that landscape by NGS, the map would certainly have some white areas. In other words: some genomic regions cannot be covered very well by sequencing the DNA with NGS technology. Here, the most important reasons for this will be explained.
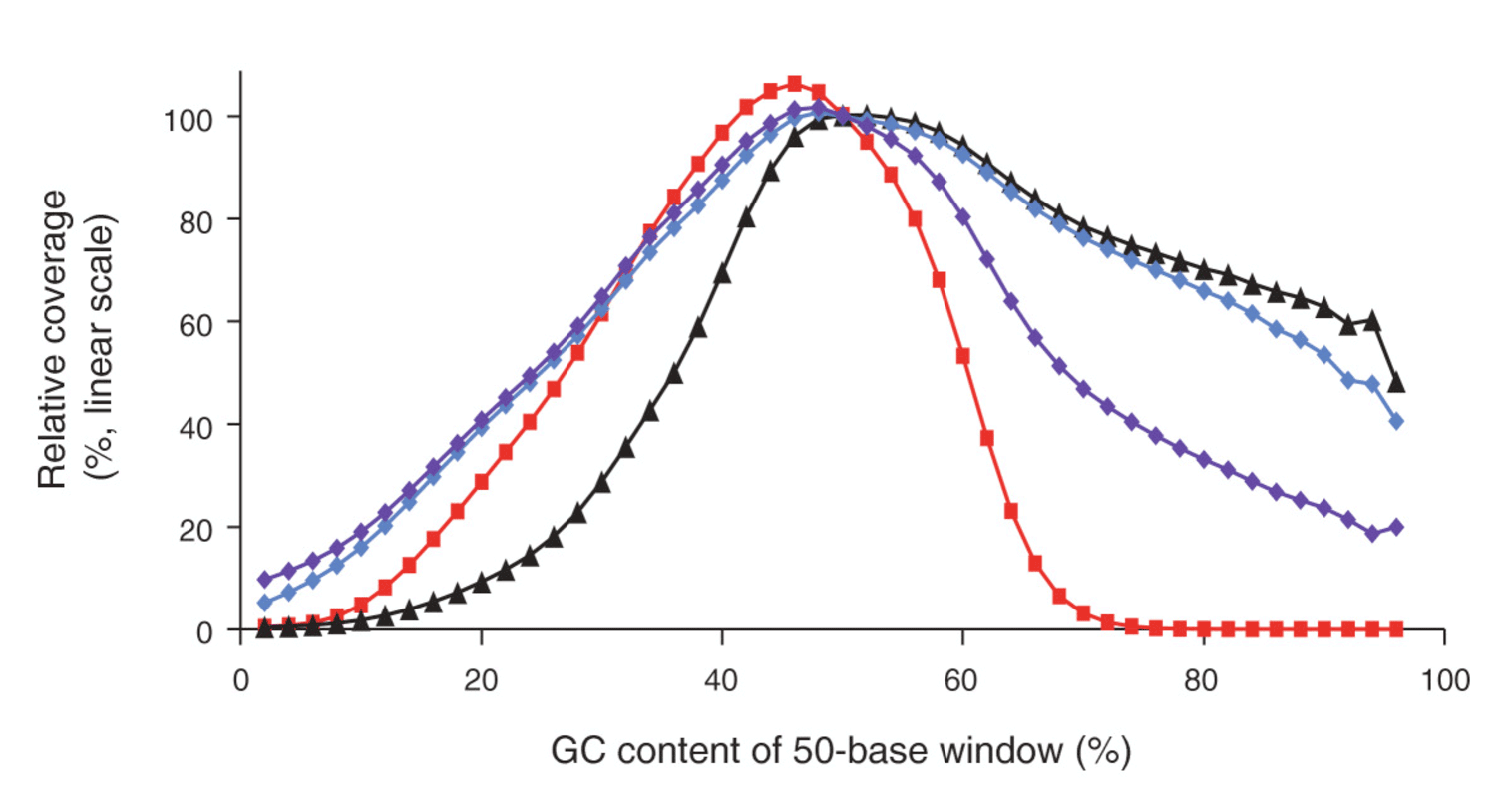


As this topic can be of high interest to bioinformatics community in general, David, could you please explain more on different platform biases and their origin? Can you also add a bit on abilities of BS-seq to solve part of this problem?
Repeats are covered by reads. They are not uncovered regions. Do you have concrete examples of uncovered regions caused by fragmentation biases? I heard people talking about depth variations caused by fragmentation biases, but my general feeling is that unlike GC biases, they are not strong enough to cause uncovered regions in long range. Another important class of uncovered regions is caused by large deletions (or equivalently, rare insertions in the reference genome). Furthermore, when you do single-cell sequencing, allele dropout probably has a much bigger effect than all the other factors combined.