Dear BioStar,
I have a bunch of sequences from Ensembl, with FASTA definition lines like so:
>ENSP00000366697 pep:known chromosome:GRCh37:9:67926761:67969840:1 gene:ENSG00000196774 transcript:ENST00000377477
When I BLAST this sequence in Amigo it returns a record that has the transcript ID as one of its external references and that shows a GO term association:
So now I'm looking for that black box where I can stick a transcript ID ("ENST00000377477") into it, and out come the GO term identifiers ("0005886", in this case). Is there an association file? It would be ideal if I can grep against such a file, because I have quite a few of these transcript IDs.
Thanks!

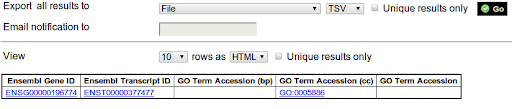

Very similar to a previous question How Do I Do Simple Go Term Lookup Given A Gene (Or Mrna) Identifier?