Greetings,
I would like to share a small tool I developed during my graduate work over the past couple years. The software is called "labnote" and is a simple python-based tool for scanning a Unix-based directory structure and generating a HTML table of contents page for various research outputs contained within those directories.
The motivation for the work is to provide a way to automatically generate a single landing page to link to outputs from things like Knitr/R Markdown and Jupyter Notebook.
In short, it helps you go from something like this:
├── co-expression
│ ├── coex-clustering-comparison
│ │ └── README.html
│ ├── hsapiens-coex-network
│ │ └── README.html
│ └── lmajor-coex-network
│ └── README.html
├── exploratory_data_analysis
│ ├── biplot_eda.ipynb
│ └── nmf_comparison.py
└── host-pathogen_networks
└── hpi-module-network
└── hpi_network_construction.py
To something like this:
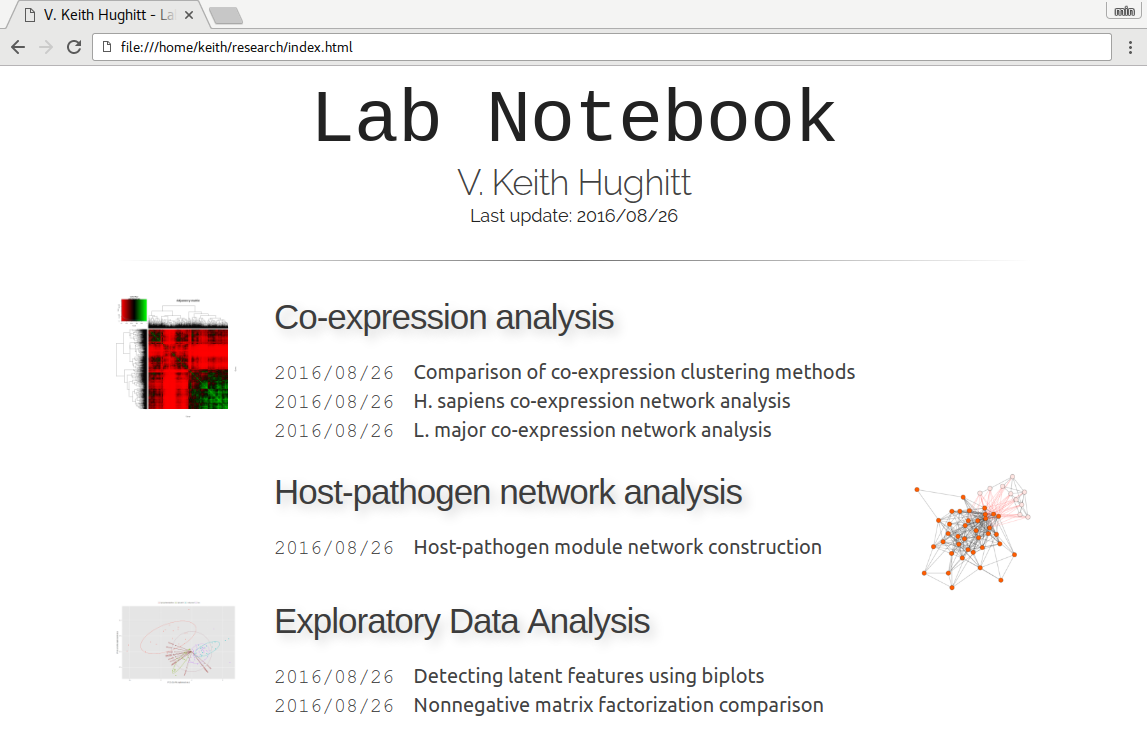
It is by no-means a fully-featured lab notebook and is missing some important features like support for multiple output versions, but hopefully some people will find it useful for their own work.
For more information on how to install and use labnote, checkout the github page at:
https://github.com/khughitt/labnote
Feedback and suggestions are welcome, as are contributions!
Cheers,
Keith

