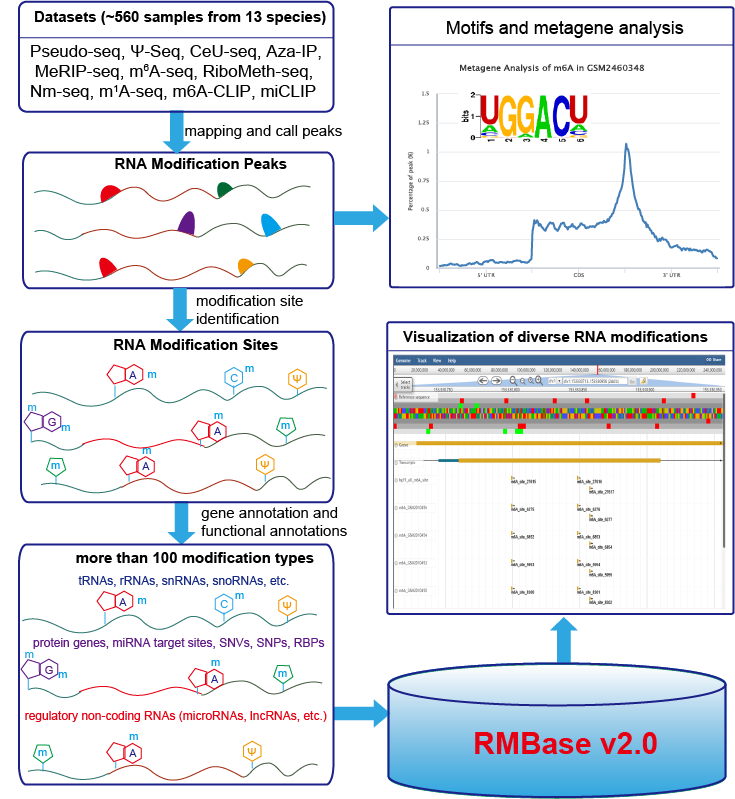
We have developed RMBase v2.0 to decode the map of RNA modifications (>100 different modification types) , as well as their relationships with microRNA binding events, disease-related SNPs or variations and RNA-binding proteins (RBP).
RMBase v2.0 was expanded with ~1402760 modification sites identified from 566 high-throughput epitranscriptome sequencing data (m1A-seq, Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, miCLIP, m6A-CLIP, RiboMeth-seq, Nm-seq) and public modification databases. It represented about 10 times expansion when compared to the previous release.
It contains ~1 379 400 N6-Methyladenosines (m6A), ~4 900 N1-Methyladenosines (m1A), ~9 600 pseudouridine (Ψ) modifications, ~1 000 5-methylcytosine (m5C) modifications, ~5 100 2′-O-methylations (2′-O-Me), and ~2 800 modifications of 100 other types.
We developed“Motif”module to provide both de novo identified position weight matrices (PWMs) and visualized logos of modification motifs.
We developed“modRBP” module to analyze the relationships of RNA modifications and RNA-binding Protein (RBP).
We developed a novel web-based tool named "modMetagene" for plotting metagenes of RNA modifications along a transcript model.
We developed modSNV and modSNP to decode the relationships between RNA modifications (m6A, m5C, 2′-O-Me) and variations (SNV, SNP), and link them to human cancers and other diseases.
RMBase v2.0 is freely available at http://rna.sysu.edu.cn/rmbase/.