Entering edit mode
7.3 years ago
biomagician
▴
410
Hi,
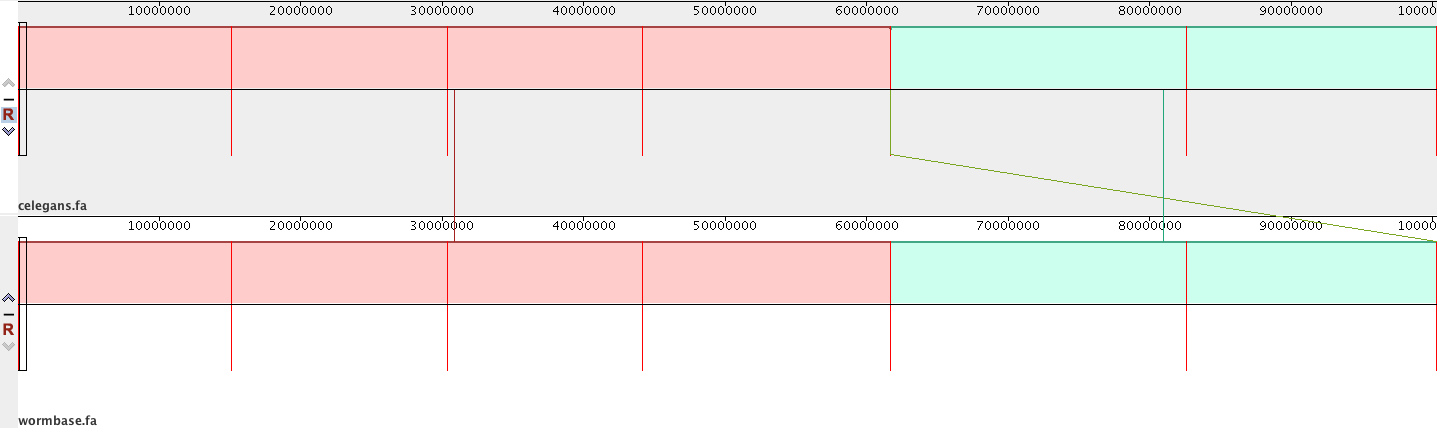
I have aligned two versions of the C. elegans genome together with Mauve. I am trying to understand how it works. Why are the blocks shown in different colours? Do I have two blocks of homology here? Can one block encompass more than one genome? What is the green line in the middle indicating? Are the red bars separating different sequences (chromosomes) in each genome?
I have exported the SNPs from this alignment and the file is empty except for the headers. Can I assume that the two genomes are exactly the same?
Thanks.
Best, C.


Different coloured blocks usually relate to separate sequences (e.g. you'd have many if you had multiple contigs). Do you know whether you should have a single complete sequence or not?
Well, there are 6 chromosomes in each file I aligned. At the bottom of the Mauve GUI I can even see on which chromosome I am and it goes from I, through II, III, IV, V to VI. That is why I do not understand why I only have 2 colours. C.