Basically, I need to perform the following tasks programmatically,
- make a csv of the parsed xml output from the ncbi blast servers programmatically;
- put data from the alignment and description in the same row, but the generators for them seem to be separate.
The safe way to do this is to Match the title of each alignment to the title of each description and then continue from there.
But, I noticed that the order in which the alignment and description appear is same.
If I used this property instead of the earlier proposed method, then it would result in a more efficient program.
I tried to verify this on a small scale ~200 alignments using a little bit of python.
blast_record = NCBIXML.parse(open('out.xml'))
for query in blast_record:
if len(list(query.descriptions)) != len(list(query.alignments)):
print(':(')
for description, alignment in zip(query.descriptions, query.alignments):
if description.title != alignment.title:
print(':(')
But If someone can really confirm that I can rely on this observation in a production environment, that would be great!
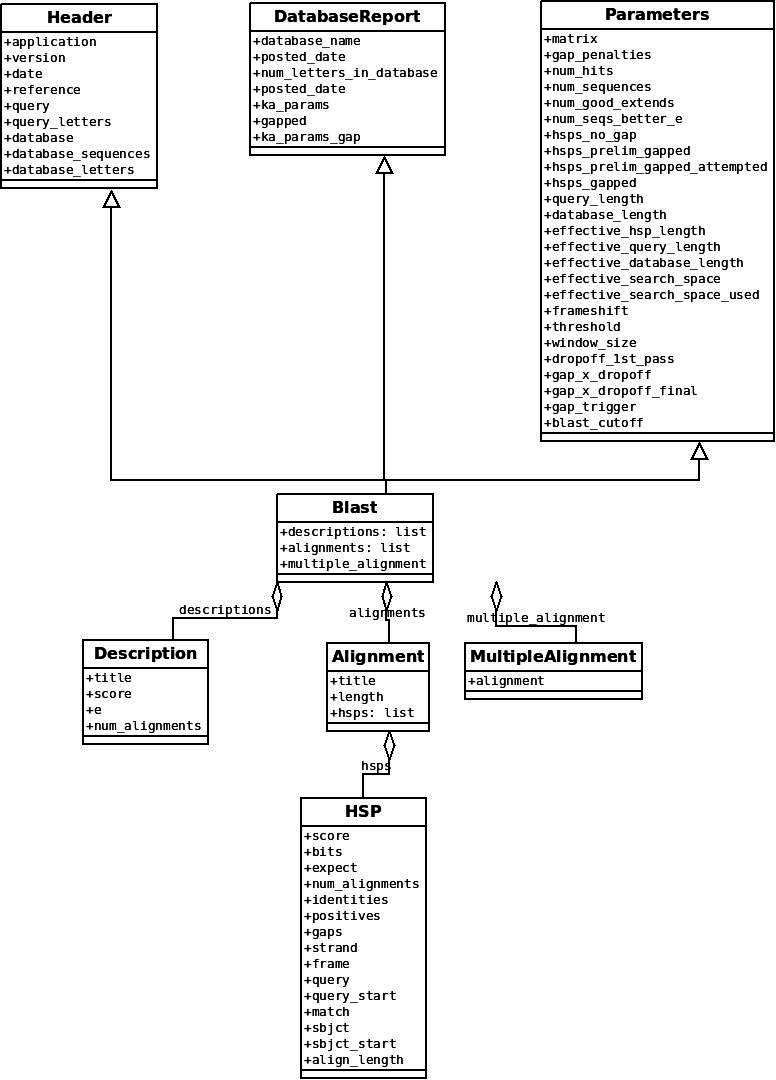
I am referring this diagram from section 7.4 of the biopython documentation