Is there any stand alone dot plot program which is like web-based in 'Plant Genome Duplication Database' or CoGE? Maybe dotter is a candidate,but I don't like its interface.
ps:It required whole genome pep blastp hit based plot,not sequence alignment based.

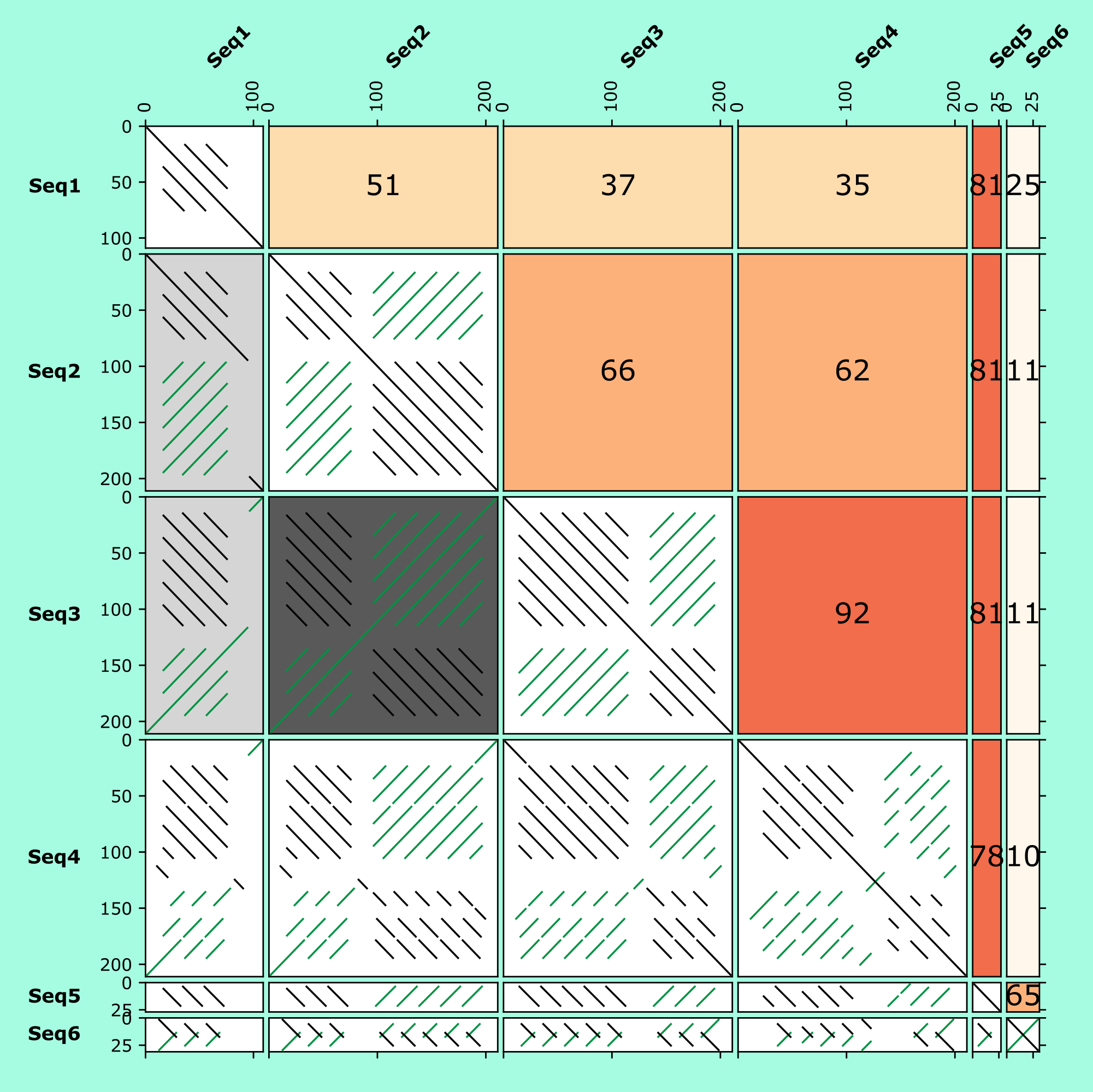

Orz,It's very very helpful.And if I want to show MCscan pairwise alignment result rather than a fast synteny scan,what could I do? Or just to transform the MCscan result to blast m8 like format ,is also can meet my job's requirement
It's very very helpful.And if I want to show MCscan pairwise alignment result rather than a fast synteny scan,what could I do? Or just to transform the MCscan result to blast m8 like format ,also can meet my job's requirement,thanks very much.谢谢
Readme of quota-alignment make me understand more clearly:)