I am interested in matching RNA motifs (Position Weight Matrices) to cDNA sequences but am confused on the correct order of operations.
I am using the R package Biostrings which only takes DNA sequences. I downloaded cDNA sequences from Ensembl.
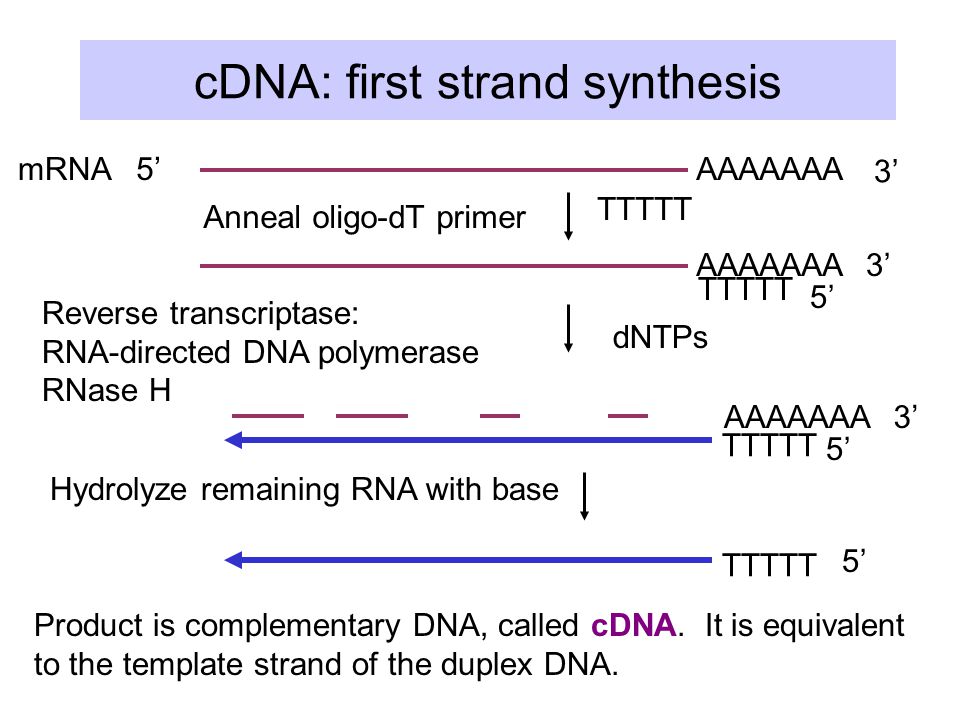 As i understand it the cDNA sequence is the 1st strand cDNA (ie: equivalent to the template strand of the genomic DNA and a reverse complement of the mRNA sequence)
As i understand it the cDNA sequence is the 1st strand cDNA (ie: equivalent to the template strand of the genomic DNA and a reverse complement of the mRNA sequence)
If my understanding is correct, since the RNA sequence is the reverse complement of the cDNA sequence then i should complement my RNA motifs into DNA (A -> T, C -> G, G -> C, U -> A) then reverse them ( reverse the column ordering of the PWM).
Which should mean my RNA_motif has been converted into a cDNA_motif which i can simply match against the cDNA sequence?
R code example:
## RNA motif "CCAU" and cDNA sequence "ATGG"
motif = matrix(c(0,1,0,0,0,1,0,0,1,0,0,0,0,0,0,1), nrow = 4)
rownames(motif) = c("A","C","G","U")
motif
# [,1] [,2] [,3] [,4]
# A 0 0 1 0
# C 1 1 0 0
# G 0 0 0 0
# U 0 0 0 1
# complement then reverse to get cDNA
rownames(motif) = c("T","G","C","A")
motif = motif[ ,ncol(motif):1]
## reorder rows for consistency
motif = motif[sort(rownames(motif)), ]
motif
# [,1] [,2] [,3] [,4]
# A 1 0 0 0
# C 0 0 0 0
# G 0 0 1 1
# T 0 1 0 0
Biostrings::countPWM(motif, "ATGG")
#[1] 1

