Hi,
I have used gffcompare to look at the transcripts in my data set and some of them are coming out with 'NA' for a class code. eg. the transcript is present when I create a transcript count matrix but when I pull out all of the class codes for my transcripts from gffcompare and merge this to the count matrix a few do not have any class code affiliated to them.
Could anyone explain how this would happen?


Please paste your code and any warning or error messages.
There aren't any errors or warning messages. My understanding is that you can use gffcompare to compare your transcripts with a reference annotation and each transcript is given a class code depending where it resides with respect to the reference annotation. I don't quite understand why some are getting no class code at all and wondered if anyone else had seen this and had a theory as to why it would happen. One explanation could be that the transcript doesn't satisfy any class but when looking at all of the class codes it looks as though every scenario is covered?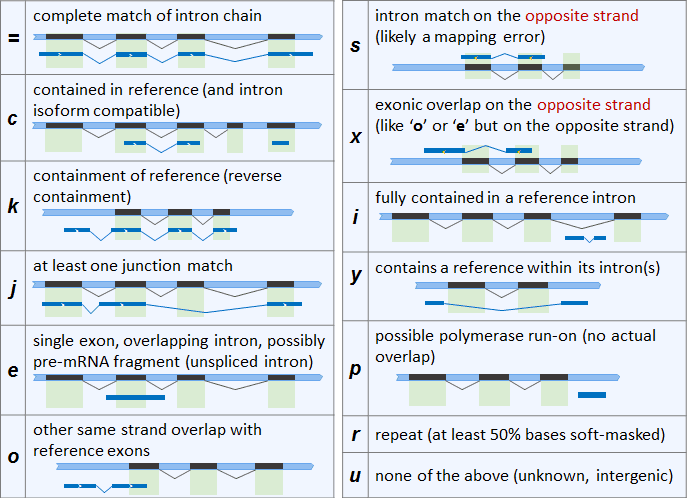
Thanks for posting more information!
I would imagine that gffcompare would assign a 'NA', also, if the contig IDs did not match between your GTF and your counts file / aligned BAM, for whatever reason. This is usually down to 'chr1' versus '1', for example.