Entering edit mode
6.8 years ago
rimjhim.roy.ch
▴
80
I have a gff file where I have already removed nested features, similar to the 1st, 2nd, 4th and 5th bed features as shown in the figure:
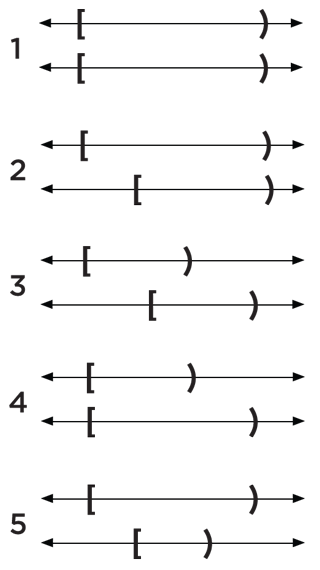
But now I want to remove features corresponding to 3, but in a way that I can keep the leftmost unique region of the first feature and rightmost unique region of the second feature, keeping the overlap as a part of the longer feature.
Example data:
scaffold1 RepeatMasker similarity 1627986 1629296 11.5 + . Clust2783_Helitron
scaffold1 RepeatMasker similarity 1628280 1638525 0 + . Clust1896_LTRRT
scaffold1 RepeatMasker similarity 1634325 1644243 0 + . Clust1098_Helitron
scaffold1 RepeatMasker similarity 1643445 1644561 2.3 + . Clust305_Helitron
Output:
scaffold1 RepeatMasker similarity 1627986 1628279 11.5 + . Clust2783_Helitron
scaffold1 RepeatMasker similarity 1628280 1638525 0 + . Clust1896_LTRRT
scaffold1 RepeatMasker similarity 1638526 1644243 0 + . Clust1098_Helitron
scaffold1 RepeatMasker similarity 1644244 1644561 2.3 + . Clust305_Helitron
Is there a simple way to do this. Please let me know if you have any further suggestions on how to remove such overlaps.
Thanks

