hi all, just as we use FASTQC for quality control check on output FASTQ files, i wanted to know if there any such commonly used pipelines/ tools for ascertaining the quality of generated VCF for downstream reporting and possibly in a clinical context.
any help from the community is much appreciated.

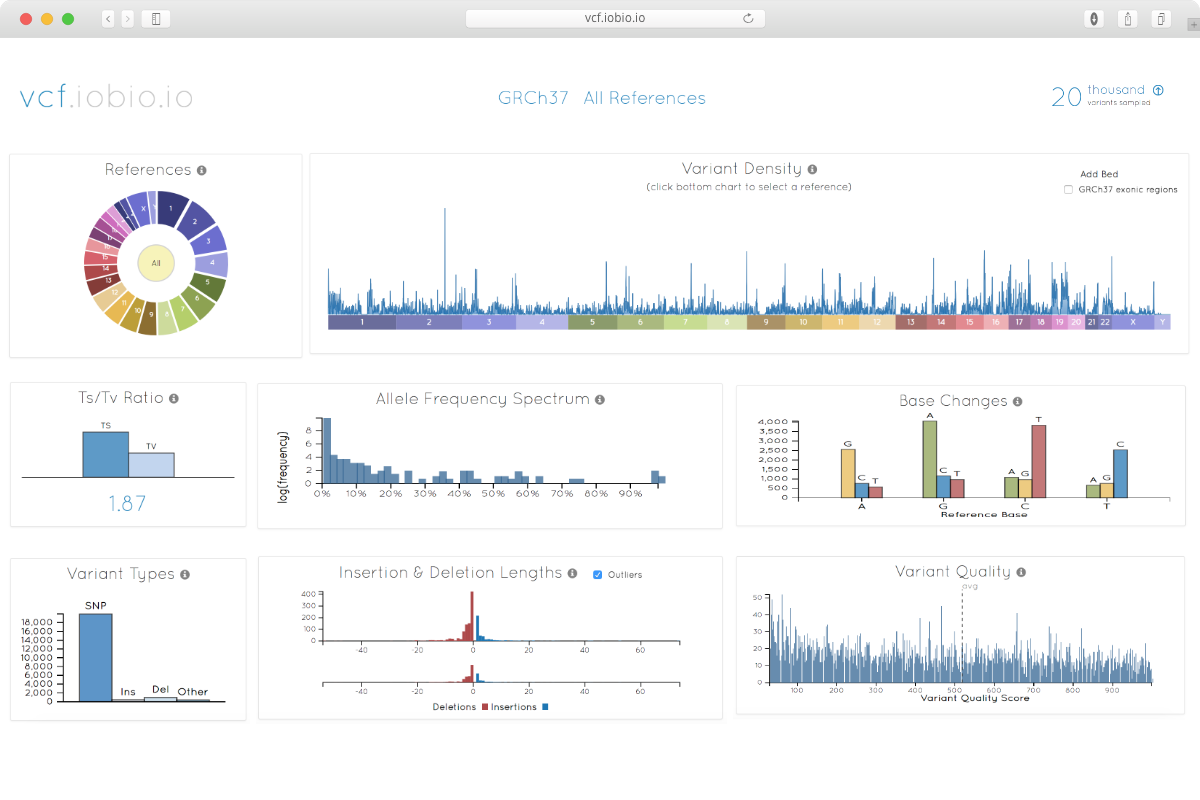

Hi,
Are you looking for format checking as header and correct field or metrics like number of INS/DEL and SNV ect ?
Hi, i am looking for metrics......
Don't know about tools , it will depends on you target (size and gene uniqueness in the genome) your organism etc...
You can include some metrics like mutational spectrum mutation rates to your calculation.
Yes, there are no standards, unfortunately. The official guidelines for reporting variants in the clinical setting in the USA and UK more or less state that ecah laboratory must define it's own QC criteria but that their method must be validated against a gold standard method, like Sanger sequencing.
For further information, you may refer to the ACMG (USA) and ACGS / NEQAS (UK / Europe). I see that you are in Inia. I do not know of regulatory bodies there, but I can put you in touch with an Indian company owner working in the area of clinical genetics (I believe).
Kevin