Hi. I finished calling SNP from genome resequencing data by using samtools. Now I need to know where the SNP located(5'-UTR, exon, intron, 3'UTR, et al). So is there any softwere I can use to build a index between genome and annotated mRNA so that I can calculate the variation of SNP in genes?
Using your SNP coordinates, if your genome is in Ensembl, you could use the Ensembl's variant effect predictor (VEP) tool: http://www.ensembl.org/tools.html
Since you are working on grape, you can use http://plants.ensembl.org/Vitis_vinifera/Info/Index
use bedtools: to know/map the SNPs to the genic regions
Simple example can be:
intersectBed -a snp.bed -b exons.bed > snpsInExons.bed
you can get the gene/exon or other information from ucsc:
ADDED:
For grape genome look annotation files;
you can extract required information from the GFF file provided.
In www.ensemblgenomes.org, go to Ensembl Plants, and choose your species from the drop-down box or "View list of all Ensembl Plants species":
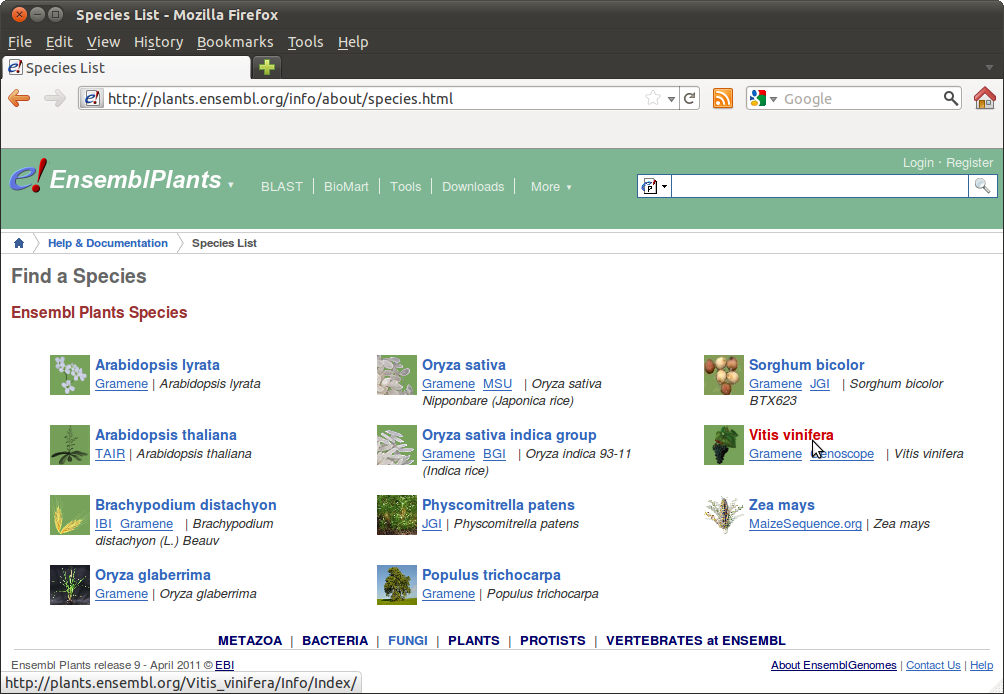
Then go to "Manage your data" and choose "Variant Effect Predictor":
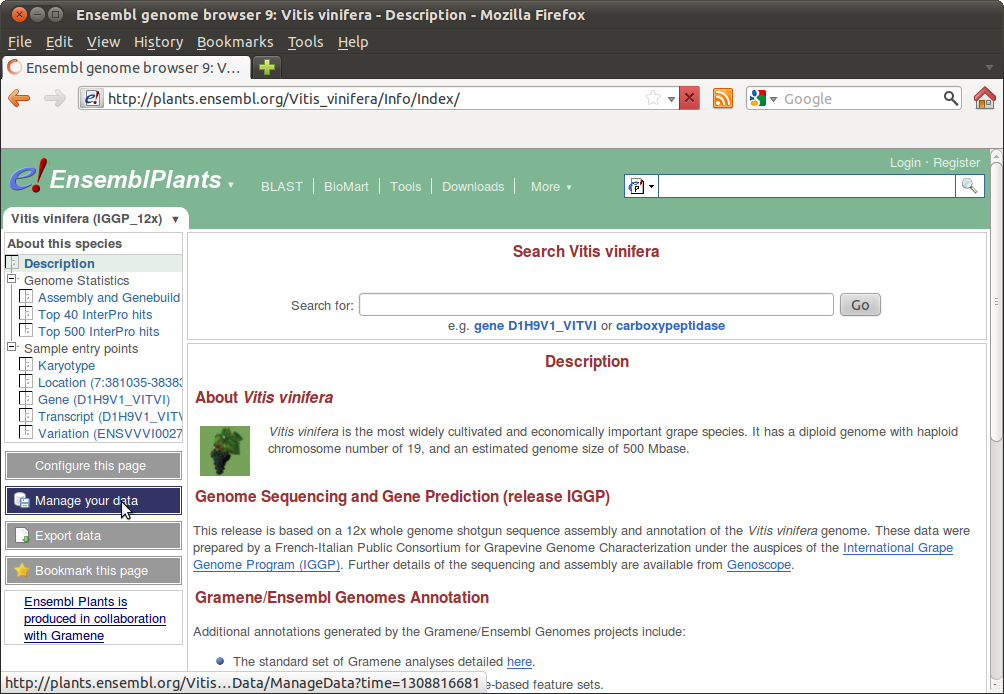
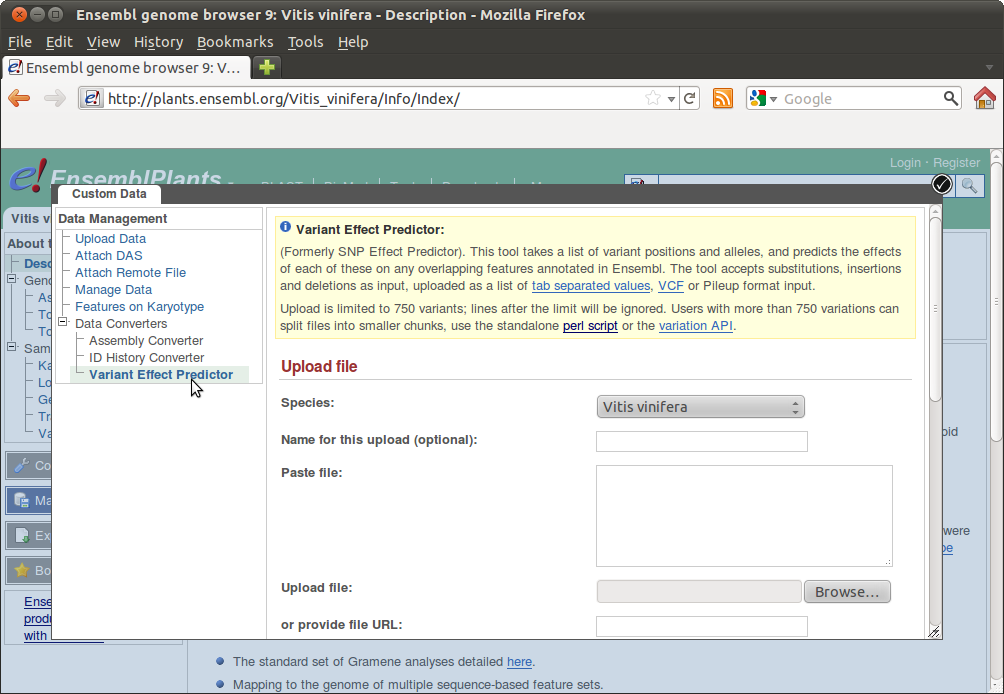
You can now copy+paste your SNPs into the form or upload your SNPs file. You can also use the Perl script for processing large batches:
Thanks for your explanation step by step. Now I know how to use it. Acturlly I am just analysis subset of genes and not the whole genomes, so that would be more convenient if I can do the idex myself by using the DNA and mRNAs for genes I analysised.It seems that I can extract the informations I needed from .GFF. I will try to do it. Thanks for all your comments.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


Ok. Thanks. I never found it before. i will try to use VEP tools. Thanks a lot for you help.
Thanks for you reply. Since I worked on grape genomes, there is no information for grape on Ensembl's. I do hava the grape genomes and predicted fasta mRNAs.So is there any softwere that can output a .gff files by using genome and mRNAs? Thanks again!