I have a question about microarray analysis,
I do the microarray analysis with the CLC Main Workbench, in the statistical section and ANOVA analysis, we have to apply filtration by FDR and Fold change to detect significant genes in an experiment.
The FDR option is available in the menu, but for fold change, I select "difference" option in experiment table to extract significant gene list.
Is the choice correct?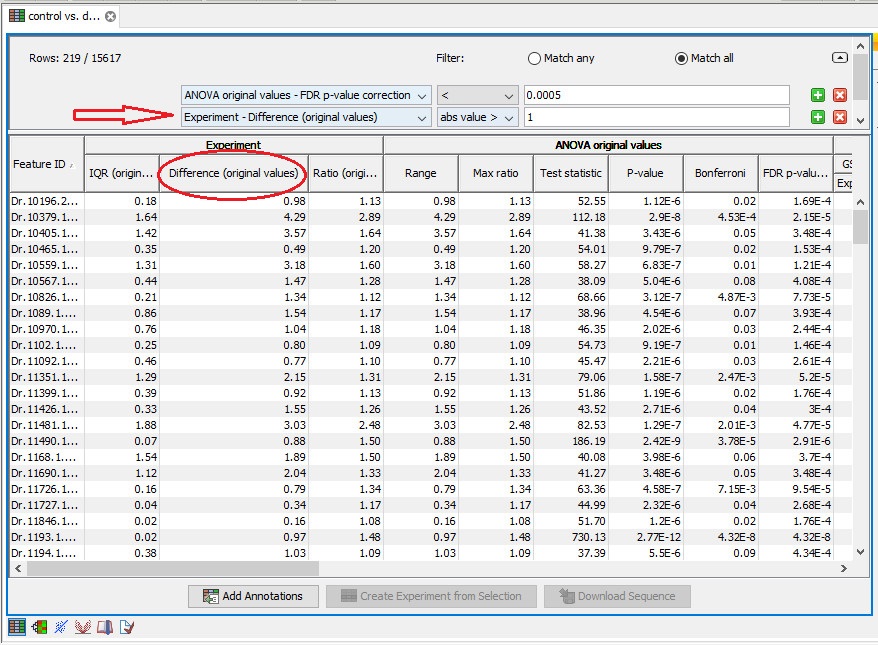
You don't have to apply fold-change filtering to detect significant differentially expressed genes - it may be interesting to do so, but you don't necessarily have to.
CLC Main Workbench version 7.8 manual has a section about "Microarray-based Expression Analysis" (chapter 4, pages 120-136), and a section "Expression analysis" (chapter 26, pages 602-655). For CLC Main Workbench version 8.0.1 manual, there is only the "Expression analysis" section (chapter 23, pages 467-524). The manual is quite detailed in both cases, if you plan to stick to CLC Main Workbench, it is worth reading it. Both manuals have a paragraph about which column to choose in case you want to filter on fold-change:
Note! It is very common to filter features on fold change values in expression analysis and fold change values are also used in volcano plots, see section 23.5.5. There are different definitions of 'Fold Change' in the literature. The definition that is used typically depends on the original scale of the data that is analyzed. For data whose original scale is not the log scale the standard definition is the ratio of the group means [Tusher et al., 2001]. This is the value you find in the 'Fold Change' column of the experiment. However, for data whose original is the log scale, the difference of the mean expression levels is sometimes referred to as the fold change [Guo et al., 2006], and if you want to filter on fold change for these data you should filter on the values in the 'Difference' column.
As we don't the scale of your data, we can't say if your choice is correct or not.
By the way, CLC used to have an outstanding customer support (at least before being acquired by Qiagen). If you are a paying customer, I recommend you contact customer support.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


You selected
chip-seqandsoftware erroras tags, which have nothing to do with your question. Please select more meaningful tags, these help people to find your question.