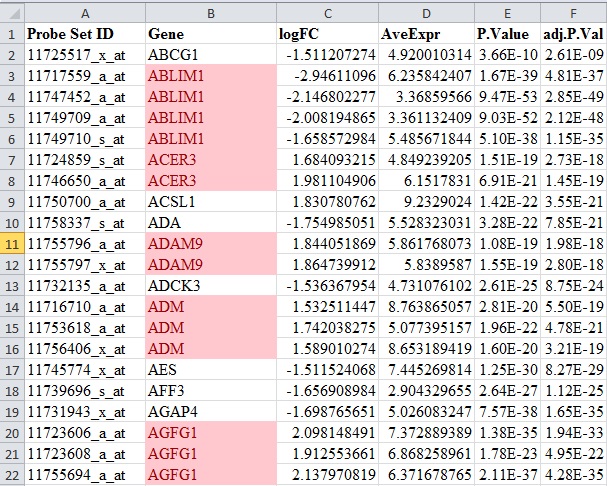
Hello everyone, I would like to know if it's correct delete duplicate genes that have the value of LogFC similiar (up or down).
I understand that each one is a different probe.
and what should I do when one probe is up and the other downregulated?
Regards,


I don't see an example of a probe-level disparity in this, could you highlight it
Dear russhh,
In my example I have only duplicate genes that follow the same direction of expression, but I have already seen results of a probe-level disparity.
So my questions are: 1. When I have "duplicate genes" that follow the same expression direction for different probes, can I delete one and keep the one with the highest expression? or what should I do?