Entering edit mode
6.8 years ago
wkmustahs21
▴
30
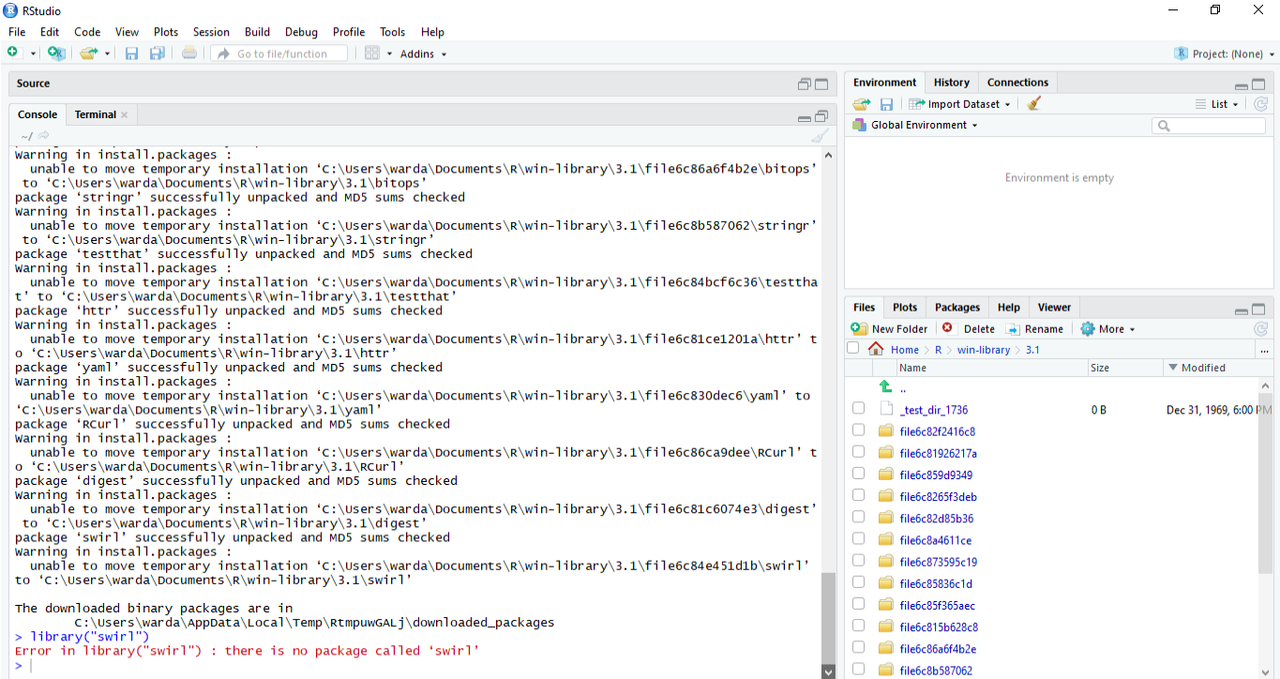
I am trying to use swirl on my R/R-studio but I am having issues with the library("swirl') part. This is what I have tried so far:
- Fixed firewall to 'Add' R gui and R-studio
- Placed myself as Admin for my personal laptop
- My R version is 3.1.3
- I have uninstalled R/R-studio several times but still nothing is really working.
So...I am not sure what to do from here to fix this issue. Does anyone have any ideas on what I can do to get this package to work??
Thanks!


How exactly did you install it?
I have just easily installed and loaded it on my R version 3.2.3 (2015-12-10) -- "Wooden Christmas-Tree". I just did:
Here is my sessionInfo....I still do not know what is causing the problem
R version 3.1.2 (2014-10-31) Platform: x86_64-w64-mingw32/x64 (64-bit)
locale: [1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages: [1] stats graphics grDevices utils datasets methods base
other attached packages: [1] BiocInstaller_1.16.5
loaded via a namespace (and not attached): [1] tools_3.1.2
Well, this is not an answer to OP issue. But do update R. It is 3.1.2 and current version is 3.5. It could be as simple as antivius + google desktop indexing as described here https://stackoverflow.com/questions/5700505/windows-7-update-packages-problem-unable-to-move-temporary-installation and https://www.r-statistics.com/2011/04/how-to-upgrade-r-on-windows-7/.
I followed the instructions here: http://swirlstats.com/students.html
I also tried your method and I am still getting the same error message.
Apart from anything else, have you tried the other suggestions by russhh and cpad? Also, have you tried to literally restart your computer and then install the package as the very first thing that you do when you go back into R? Install like this (not via install.packages):
I'm fairly certain
swirlis CRAN not bioconductor. You really _should_ be installing it withinstall.packagesThat's funny, russhh, because I got it working via Bioconductor and was even playing around with the package yesterday. Conversely, on so many occasions, installing via
install.packagesfrom CRAN has led to issues with version number incompatibilities between the installed R version and the correct package version to obtain.I stand by my advice to try to install via Bioconductor
You could define a writeable directory for storing your installed packages. For example, you could set R_LIBS_USER as described in this stack overflow post (the second answer, not the accepted answer). It seems extremely weird that you can't write to your own documents directory though.
also weird is the timestamp on the file down right corner on a single file. Currently OP is in temporary folder, not in user folder.
That is unix epoch time.
Good spot, cpad. How many of us were even alive in 1969?
Probably as separate x and y chromosomes
hi russhh
Would I make this directory in this line of script: trace(utils:::unpackPkgZip, edit=TRUE)?
No, you don't do that in R, you should follow the instructions in the stack overflow post: it explictly tells you how to set environment variables.