So I have my time series RNA-seq data, which I conducted some k-means clustering on in R to have a look at how it clusters. I am wondering if there are any good tools in R to then analyse the genes within each cluster, or the next step from the k-means clustering? Each cluster has around 50 genes in them.
I've been thinking about investigating nearby transcription factors (mouse cells) however the closest R tool I can find for doing that is pwOmics. I know that the TFcheckpoint webtool is a thing, however I was hoping to avoid copy-pasting forty different sets of genes in there from text files. Are there any similar R tools?
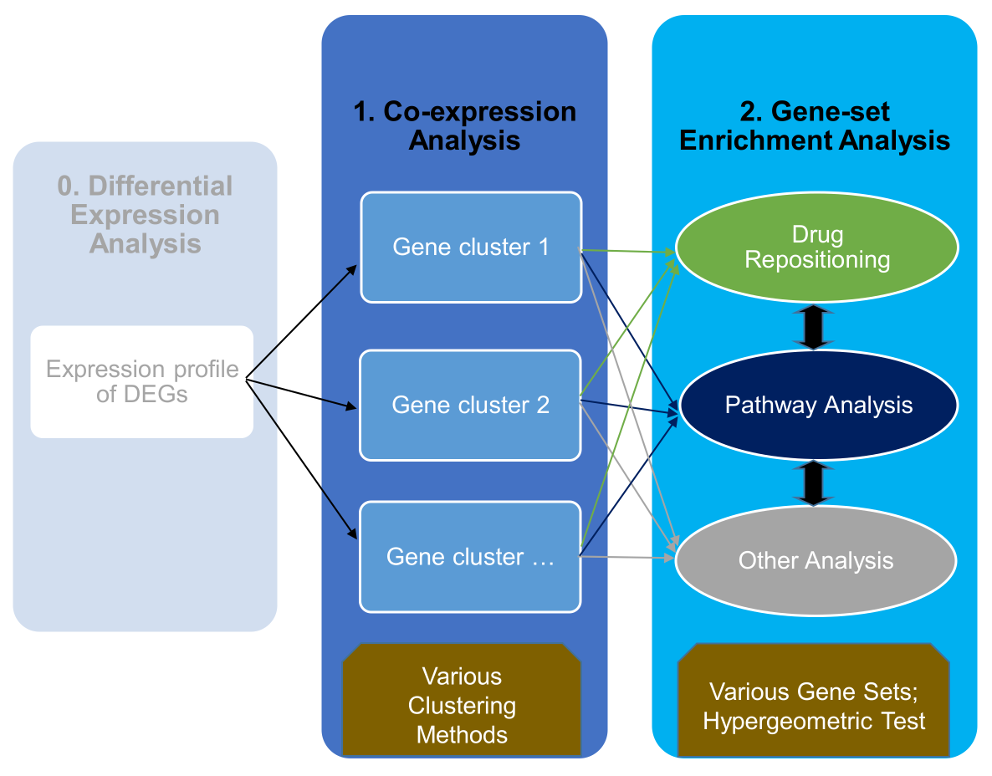
The main thing is that I am a bit unsure where to go from here, as there are a lot of genes within each cluster, and I have about four different plots with about 10 clusters in each. Any suggestions would be super useful! Thanks

 .
. and
and 

Let's say each cluster is one gene set. Taking each of this gene set you could do GO and pathway analysis to understand the biology. Simplest way to do this is DAVID. Further downstream analysis depends on what question you want to address.
Annotation enrichment analysis is a typical way of looking at clusters of genes. There are different R packages for this, e.g. Bioconductor topGO for GO terms enrichment or, in simple situations, you could do it with the fisher.test() function.
As an aside, you may want to have a look at this post about clustering time series.