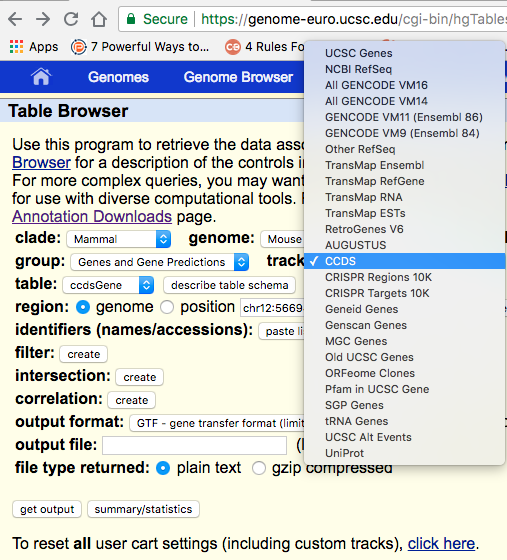
Hi all, I prepare to download annotation file(gtf) from ucsc to do rna-seq analysis, but i can not download directly. So i preapare to download mouse txt file and use genePredToGtf to transfer it to gtf file. I have a question, the table browser page has "track:" ,it has different choice. like ucsc genes sgp genes and so on. which one should i choose? Thanks!
0
Entering edit mode
6.7 years ago
Denise CS
★
5.2k
Tracks are different annotations that can be added to your view in the UCSC genome browser; so it's really up to you which one to choose. They are aligned to the genome and can be either computed by UCSC or provided by others.
Do you want the ab initio prediction only? Then, choose AUGUSTUS. Are you interested in a set of annotations of the coding genome that is an agreement between several resources? Then choose CCDS. And so on so forth.
Selecting the "output format" as GTF and then hitting "get output" works for me:
Similar Posts
Error! readyState=0 status=error text=
Loading Similar Posts
Traffic: 1821 users visited in the last hour
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


I've added the tags UCSC, GTF and Table browser to your query. People on BioStars use the tags to search for questions that they might be able to answer, and follow relevant tags with My Tags, so judicious use of tags can help people to find and answer your questions. We recommend tagging with the name of the tool you are trying to use, the type of analysis you're trying to do and the file-types you're working with.
Why can't you download a GTF directly?
Thank you for add the tag for me. They have the same gene id and transcript id, which will effect the analysis later (This can lead to problems with downstream analysis tools that expect exons ofdifferent isoforms to have the same gene_id, but different transcript_ids.).
Have you tried downloading alternative GTF files, eg Ensembl?