I am having trouble adding the GO accession numbers on the left hand side of this graph. They match up with the GO descriptions found on the right hand side of the Heatmap. I thought I could use the text function, seen after the Heatmap.2 function, to annotate them, however it doesn't work.
If anyone has any suggestions to do this via R I would be very grateful!
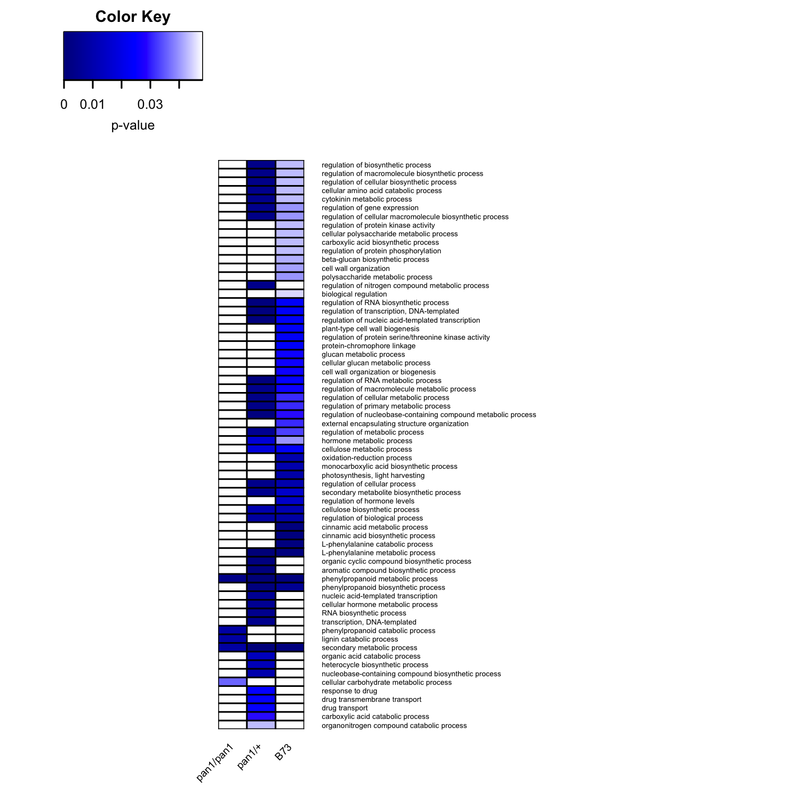
png("BPheatmaps_in_r.png", # create PNG for the heat map
width = 5*300, # 5 x 300 pixels
height = 5*300,
res = 300, # 300 pixels per inch
pointsize = 8) # smaller font size
heatmap.2(BPmat_data,
col = c(colorpanel(1,"white","white"), colorpanel(700,"darkblue","blue","white")),
margins = c(4, 28),
dendrogram = 'none',
trace = 'none',
lhei = c(2, 8), #plot layout
scale = c("none"),
symbreaks = min(BPmat_data, na.rm=TRUE),
cexRow = 0.5, cexCol = 0.7,
density.info = "none",
sepcolor = colorpanel(1,"black","black"),
sepwidth=c(0.0001,0.0001),
key.xlab = "p-value",
colsep=0:ncol(BPmat_data),
rowsep=0:nrow(BPmat_data),
srtCol=45)
get.uni <- !duplicated(BP$GO_Term)
text(x =5, y = seq(0.2,1.2, by=(1/nrow(BPrnames))),
labels = BP$GO_Term[get.uni],
las = 2, col = "#000000", cex = 0.4, xpd = TRUE)
dev.off



Please read the how to on adding images: How to add images to a Biostars post