I have seen this figure in a Nature Immunology paper. I was wondering where I can find the genotype data from the 36 populations (32 Eurasian populations and 4 HapMap populations) in panel b. Do you know if it is open access? I couldn't find anything online.
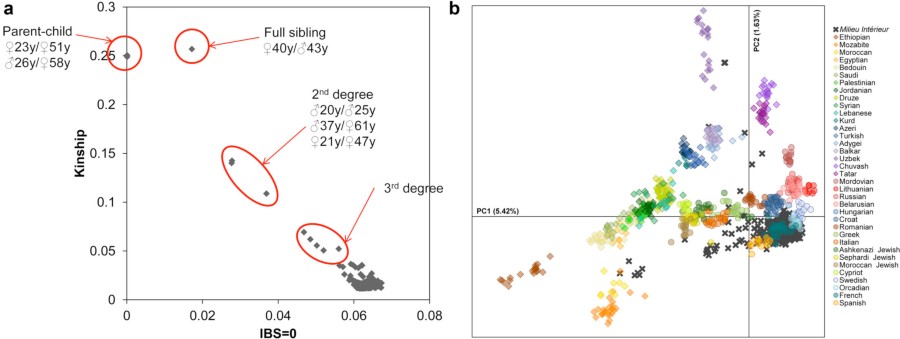


Sure, it is Supplementary Figure 21 of https://www.nature.com/articles/s41590-018-0049-7
Thanks - you should check the supplementary information for the purposes of finding out where the data is actually stored. I imagine that it is controlled access.
By the way: one project that I failed to mention was Greater Middle East