Hi,
I performed a GWAS on a BMI data set, which has ~570 individuals. I used plink to do the analysis and found some SNPs that reached suggestive or near-genome-wide significance.
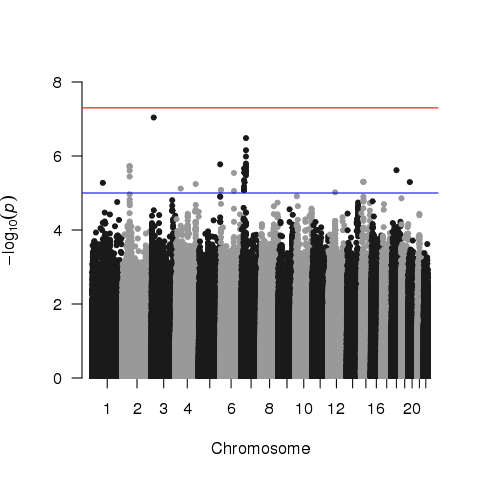
I expected these SNPs to be observed in other publications. But, most of these SNPs have never been reported before, which made me alert. I then took a look at QQ plot:
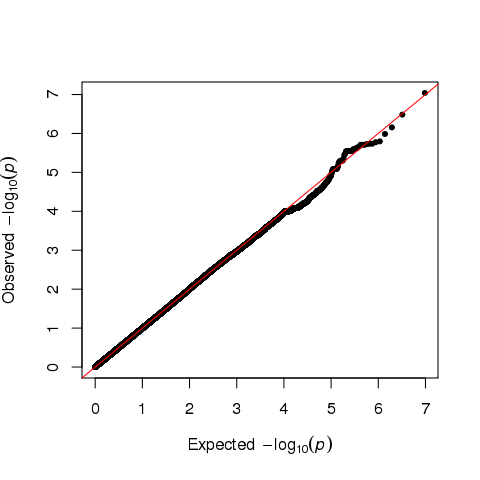
I think there is no enrichment from the QQ plot.
It is possible that the data set is small and few individuals have extreme phenotype value, so the statistical power in detecting significant SNPs are small.
But is it possible that there are some problems with my analysis?
Thanks!

