Homology means shared evolutionary ancestry. Sequence similarity is often used as a proxy for homology but inferences should be made with care.
The similarity between two genes/proteins should not just be good but has to be statistically significant (metrics like E-value) for the two genes/proteins to be considered homologous.
INFERRING HOMOLOGY FROM SIMILARITY
The concept of homology – common evolutionary ancestry – is central to
computational analyses of protein and DNA sequences, but the link
between similarity and homology is often misunderstood. We infer
homology when two sequences or structures share more similarity than
would be expected by chance; when excess similarity is observed, the
simplest explanation for that excess is that the two sequences did not
arise independently, they arose from a common ancestor. Common
ancestry explains excess similarity (other explanations require
similar structures to arise independently); thus excess similarity
implies common ancestry.
However, homologous sequences do not always share significant sequence
similarity; there are thousands of homologous protein alignments that
are not significant, but are clearly homologous based on statistically
significant structural similarity or strong sequence similarity to an
intermediate sequence. Thus, when a similarity search finds a
statistically significant match, we can confidently infer that the two
sequences are homologous; but if no statistically significant match is
found in a database, we cannot be certain that no homologs are
present.
Pearson, 2013
Members of a protein family are descendants of a common ancestor and are hence homologous. However, in the course of evolution they would have acquired new domains or reshuffled their domains such that their sequences are no longer similar. Proteins that have full length sequence similarity are called homeomorphic (Wu et al., 2004). Therefore, members of a protein family may be homologous but not homeomorphic. However, homeomorphic proteins can evolve independently and therefore may not be considered homologous.
Identifying homologous proteins is, therefore, not a simple task. Machine learning algorithms are used for better identification of homologous proteins. Some of these algorithms are mentioned in the linked papers.
In general, global similarity, rather than local similarity should be considered for identifying homeomorphs. See https://biology.stackexchange.com/q/11263/3340
I don't know the proteins in your example but if they are from same protein family, then they are homologous. As someone else pointed out, these genes are indeed paralogs.
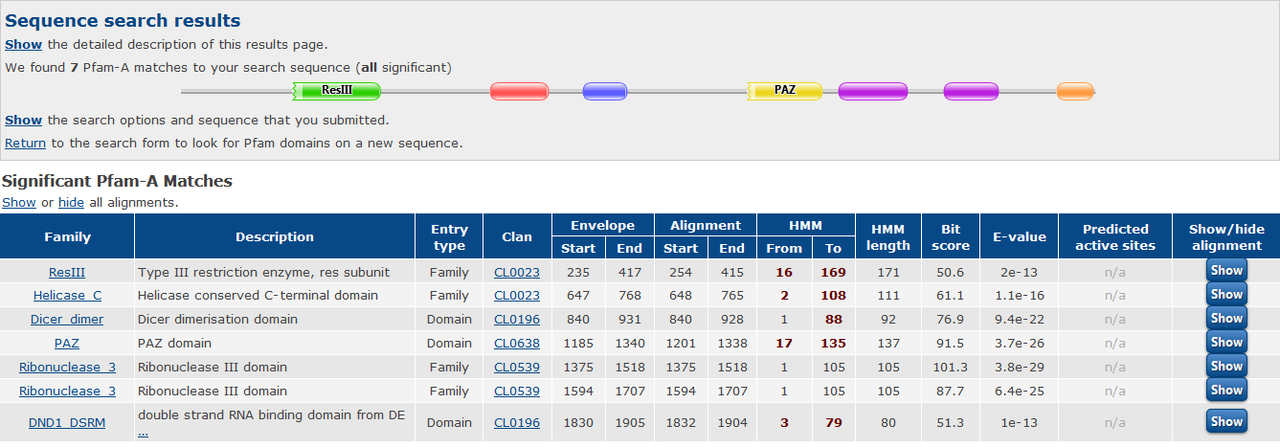
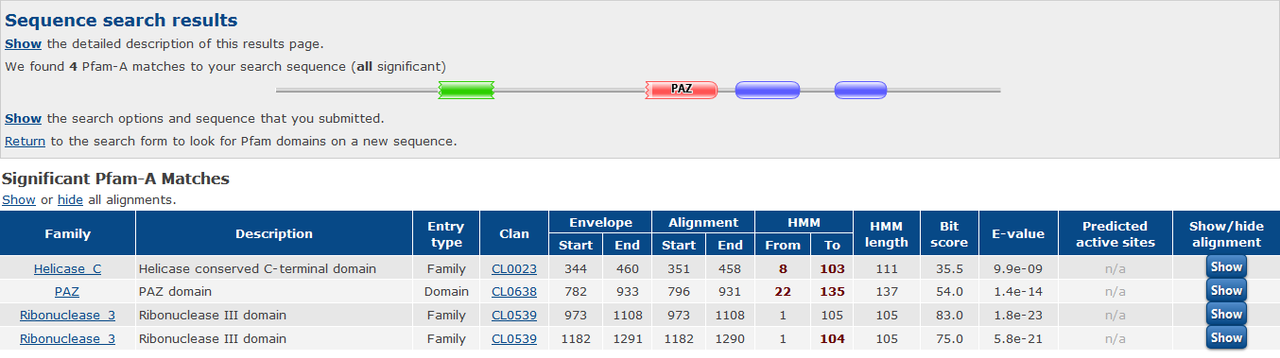


How would you define the relationship between these two sequences at the moment? Would you call them homologous? How similar are these two protein sequences?
https://en.wikipedia.org/wiki/Sequence_homology
I define the relationship based on shared domains between two genes. these two proteins have 70 to 100 percent identity in shared domains and in the rest of their sequence, those are not meaningfully similar.
Sej Modha is asking about the ancestry relationship between the sequences. Are you comparing two proteins from different species, or from the same species?
the two proteins are from Arabidopsis thaliana. AT1G01040 and AT3G43920
Two proteins can be either paralogous or orthologous, not homologous. Homologous is a super term for paralogs and orthologs. When you say, homologous it is confusing.
I'd say that's not strictly true, the proteins are/can be homologs. They do not 'share some homology', as it's an absolute term, which is where a lot of confusion arises. But whether they are paralogs or orthologs, they are both still homologs. I agree its less clear-cut, but to say that they can't be homologous is misleading IMHO.
Then, these two proteins are homolog or not? or maybe are homolog in some domains? :)
I am still confused about the following statement:
It is not clear whether you'd like to state that these proteins are homologs of protein X or simply that these two proteins have certain shared regions i.e. coding for the same domain.
Sorry for poor English. I want to know that, based on the above pfam results, are these two proteins homolog of each other or not?
Based only on that Pfam result, the answer is probably, but you can't really say for certain. There are a lot of caveats. h.mon's answer below is the correct one as this has already been previously determined.
Looking at the screenshot of some colored boxes is not enough to decide if proteins are homologous or not. Check the genes of interest at the pATsi database.
http://cab.unina.it/athparalog/main2.html
I know about paralogy and orthology, duplication and speciation. I used the super term "homology" to avoid talking about differences between paralogs and orthologs. to be more clear, both these genes are in arabidopsis thaliana.
I think this is cross-posted
https://biology.stackexchange.com/questions/77050/are-these-two-proteins-homolog
Hello utsafar!
It appears that your post has been cross-posted to another site: https://biology.stackexchange.com/questions/77050/can-two-proteins-sharing-a-few-domains-be-considered-homologous
This is typically not recommended as it runs the risk of annoying people in both communities.
I believe biostars should be merged with stackexchange. Since biostars is an independent platform we cannot moderate cross-posts and migrate questions. It is certainly annoying but this problem was created by the administrators. There is no way to co-ordinate between these two communities. In fact now there is a bioinformatics stackexchange as well.
This site existed long before the Bioinformatics stack exchange. The prospect of merging has been bought up previously and never gets very far. Some of the Biostars mods are Bioinformatics SE moderators too. A happy medium might be some more moderators which have priviledges on both sites to shut cross posts down in a timely manner.
One of the reasons people like coming to Biostars, in particular, is that SE is quite a hostile place. Biostars is much more welcoming to novice users of bioinformatics tools, which off the top of my head, I'd say is close to, if not the majority of the posts we get.
In practice, I don't think we experience a significant degree of crossposting (at least not that is spotted anyway).
As pointed out by jrj.healy, biostars predates the bioinformatics stackexchange (it may well predate the biology stackexchange as well). Also, since both Heng Li and I are moderators on stackexchange (the bioinfo site, not the biology site) too, we actually can moderate things on both. Obviously posts can't be migrated between independent sites, but that's usually a non-issue...just pick a given site for a particular question.
Biostars and Biology SE are two different communities with probably different users. I cross-posted my question to use the knowledge of both communities. Even, I think I must note that I posted this question in both sites.