Entering edit mode
6.1 years ago
hirad.alipanah
▴
10
Hi everyone. We have the following table for our data:
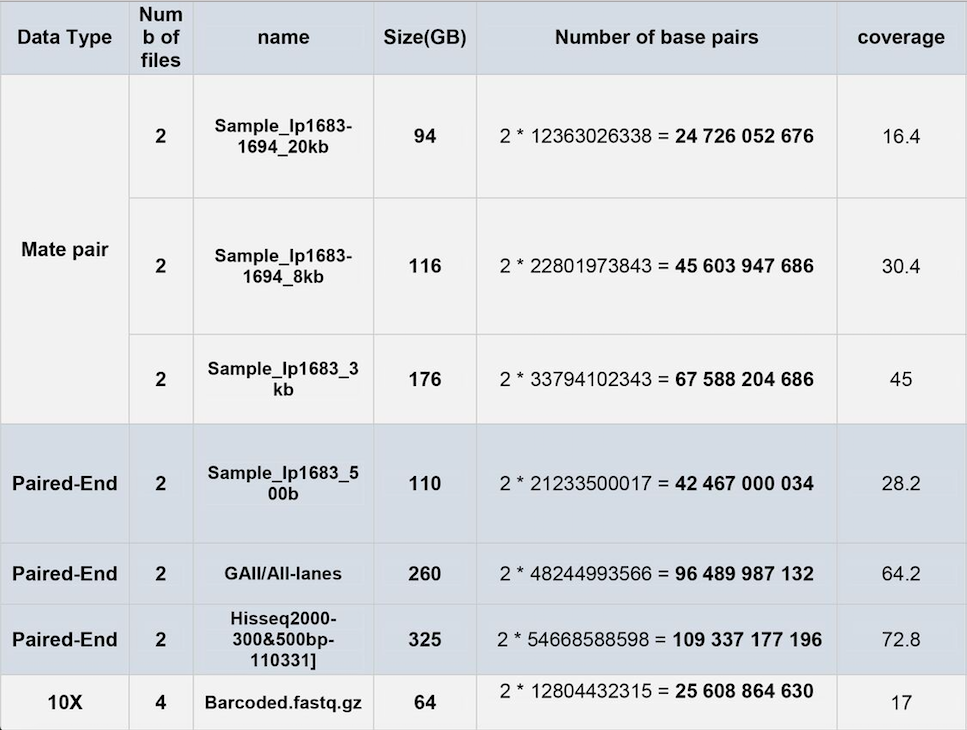
We want to assemble the genome of the planaria "dugesia japonica". Its genome is approximately 1.5Gbp, diploid and very repetitive (similar to Smed.) What is the best assembler that you suggest for assembling all of this data?


Could you clarifiy on which species you want to perform the assembly ?
Yes. the planaria "dugesia japonica"
Could you edit your question to add these information + expect size of genome + ploidy , etc... Thanks
10x data may need to be handled separately.
supernovais what you would want to use there. I see some data from GAII which would lead me to believe that you have collected different data over time. Are all these datasets for the same exact sample/organism?Yeah, that was our first option. But how do we use the output of
supernovafor our other data? Yes, the datasets are from the same exact organism. But they are from different samples.I can recommend
ABySS, very versatile, excellent cluster usage and quite performant, it might require some parameter tweaking though (as with most assembly software). From the same developers there are also tools to include the 10x data.Yes. We've considered that, too. But it needs a lot of memory (around 1TB but we only have 500GB.) Do you know other assemblers that require less memory?
perhaps
soapDeNovois an option ? (no experience with myself though).Masurcawill likely also be too mem intensive. I think in most cases you still need to figure out how to include the 10x as there are very few (to none?) software that will be able to process all your data at once.