Entering edit mode
6.3 years ago
WUSCHEL
▴
850
For downstream data analysis, how can I use tidy gather function for df with more than one header,
I want to plot a timecourse experimental data of 5 genotypes in a one bar plot like below
example df
structure(list(X1 = c("0d", "WT", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0"), X2 = c("0d", "aox2-1", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0" ), X3 = c("0d", "aox5-1", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0" ), X4 = c("0d", "aox7-2", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0" ), X5 = c("0d", "aox9-1", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0", "0" ), X6 = c("12h", "WT", "0.646", "0.632", "0.658", "0.635", "0.649", "0.692", "0.687", "0.669", "0.663", "0.681", "0.689", "0.666", "0.677", "0.664", "0.652", "0.651", "0.641", "0.657", "0.658", "0.642"), X7 = c("12h", "aox2-1", "0.653", "0.619", "0.611", "0.644", "0.597", "0.605", "0.581", "0.588", "0.624", "0.619", "0.635", "0.626", "0.625", "0.63", "0.612", "0.633", "0.636", "0.586", "0.633", "0.609"), X8 = c("12h", "aox5-1", "0.635", "0.609", "0.604", "0.601", "0.622", "0.591", "0.569", "0.585", "0.576", "0.56", "0.609", "0.583", "0.561", "0.62", "0.651", "0.587", "0.642", "0.621", "0.574", "0.573"), X9 = c("12h", "aox7-2", "0.541", "0.532", "0.566", "0.537", "0.6", "0.571", "0.6", "0.594", "0.594", "0.592", "0.516", "0.54", "0.515", "0.557", "0.607", "0.586", "0.549", "0.557", "0.531", "0.56"), X10 = c("12h", "aox9-1", "0.616", "0.608", "0.615", "0.614", "0.652", "0.641", "0.629", "0.623", "0.613", "0.607", "0.585", "0.575", "0.633", "0.632", "0.561", "0.571", "0.563", "0.62", "0.565", "0.565"), X11 = c("24h", "WT", "0.739", "0.732", "0.732", "0.72", "0.716", "0.744", "0.747", "0.726", "0.737", "0.74", "0.724", "0.73", "0.708", "0.711", "0.717", "0.739", "0.738", "0.709", "0.722", "0.752"), X12 = c("24h", "aox2-1", "0.732", "0.715", "0.707", "0.725", "0.727", "0.727", "0.728", "0.736", "0.734", "0.731", "0.713", "0.709", "0.71", "0.718", "0.738", "0.708", "0.728", "0.721", "0.72", "0.714"), X13 = c("24h", "aox5-1", "0.746", "0.735", "0.713", "0.716", "0.746", "0.728", "0.745", "0.752", "0.726", "0.713", "0.71", "0.721", "0.715", "0.713", "0.712", "0.738", "0.741", "0.737", "0.729", "0.719"), X14 = c("24h", "aox7-2", "0.706", "0.714", "0.715", "0.695", "0.696", "0.714", "0.703", "0.672", "0.677", "0.694", "0.686", "0.706", "0.724", "0.726", "0.706", "0.694", "0.709", "0.725", "0.714", "0.711"), X15 = c("24h", "aox9-2", "0.723", "0.715", "0.706", "0.702", "0.702", "0.733", "0.726", "0.732", "0.725", "0.719", "0.719", "0.711", "0.699", "0.713", "0.705", "0.732", "0.725", "0.729", "0.723", "0.721")), row.names = c(NA, -22L), class = c("tbl_df", "tbl", "data.frame"), spec = structure(list( cols = list(X1 = structure(list(), class = c("collector_character", "collector")), X2 = structure(list(), class = c("collector_character", "collector")), X3 = structure(list(), class = c("collector_character", "collector")), X4 = structure(list(), class = c("collector_character", "collector")), X5 = structure(list(), class = c("collector_character", "collector")), X6 = structure(list(), class = c("collector_character", "collector")), X7 = structure(list(), class = c("collector_character", "collector")), X8 = structure(list(), class = c("collector_character", "collector")), X9 = structure(list(), class = c("collector_character", "collector")), X10 = structure(list(), class = c("collector_character", "collector")), X11 = structure(list(), class = c("collector_character", "collector")), X12 = structure(list(), class = c("collector_character", "collector")), X13 = structure(list(), class = c("collector_character", "collector")), X14 = structure(list(), class = c("collector_character", "collector")), X15 = structure(list(), class = c("collector_character", "collector"))), default = structure(list(), class = c("collector_guess", "collector"))), class = "col_spec"))


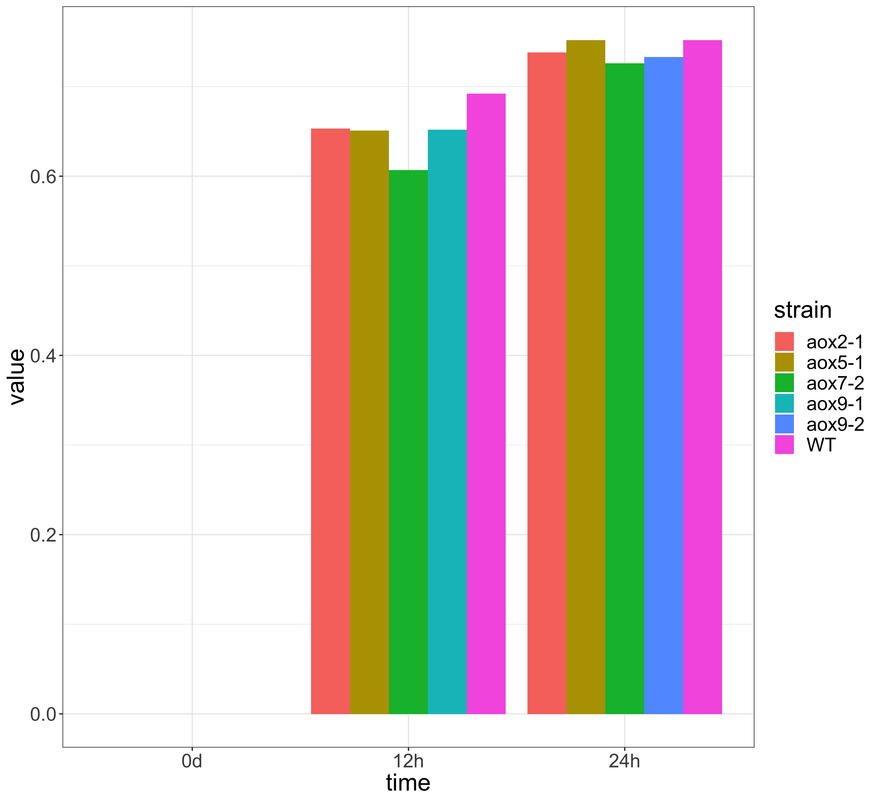

after some code cleaning:
Thanks a heap cpad0112 :) BTW, can you help me with the adding SE to the bars if possible.
Once wide format is converted to long format, use summarySE (from Rmisc librar) function to calculate SD, SE etc. Combine these values with long format data and use geom_errorbar function to plot error bars.
Hi cpad0112, don't you think using
sapply, subsetting by[]andstr_split_fixedmakes less readable compare to usingtidyverseverbsslice,summarise_allandseparate. Just a thought. Nothing personal :)sapplystep is optional, as OP may have the numeric data and I kept it out of the tidyflow. For other stuff, your suggestions are good. Next time, would incorporate that.some thing like this:
Thank you Chirag, Appreciate.