Dear all
I have extracted reads NOT mapped to the human genome and re-aligned them to another genome called 'V' that DOES NOT CONTAIN bacterial sequences. I set some regions of interest, copied the sequence and BLAST it. For instance, for this region:
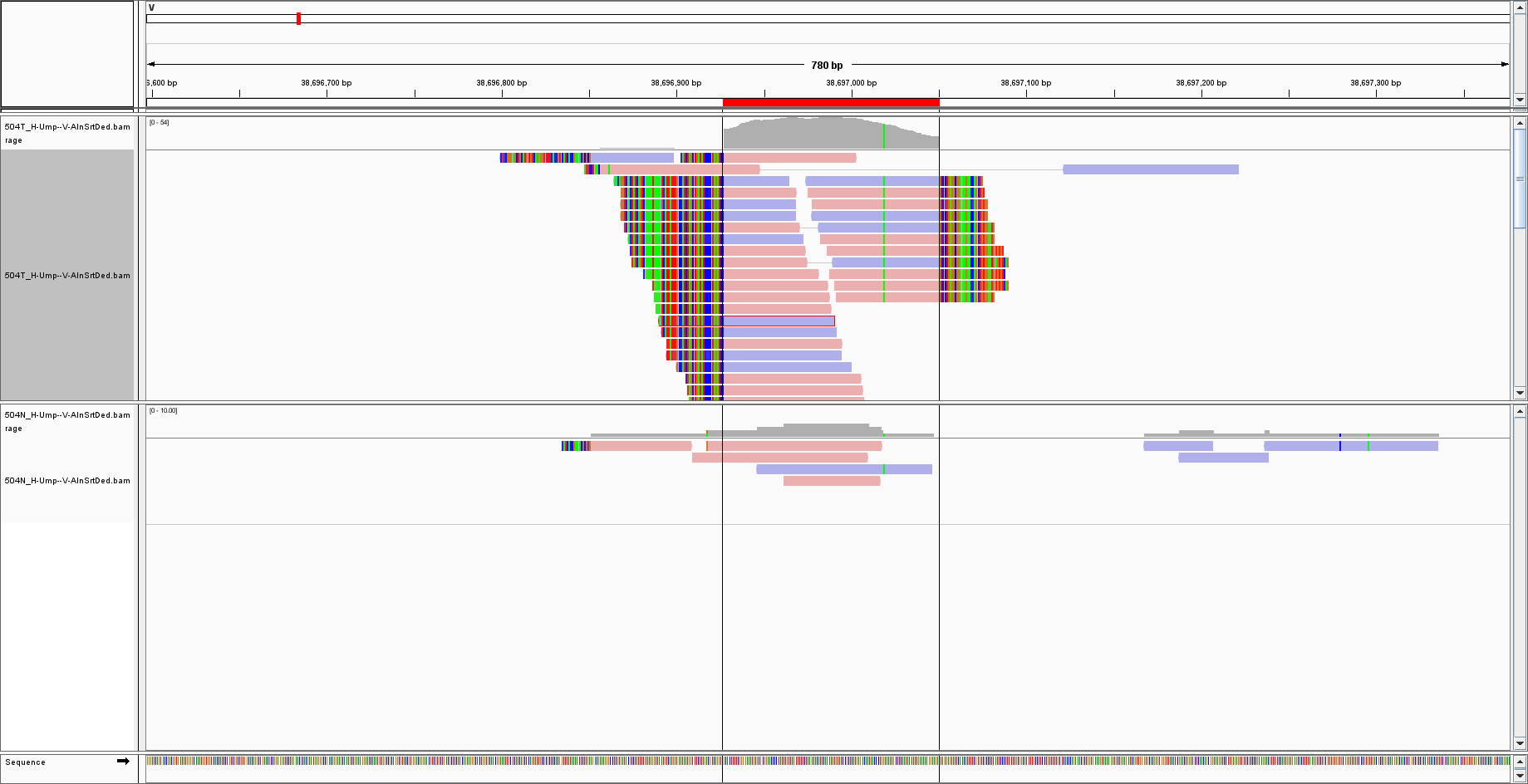 the BLAST result gives the top hits as:
the BLAST result gives the top hits as:
Escherichia coli strain 2248 plasmid pNDM-2248 (coverage 100%, e-value 1e-56)
Salmonella sp. strain Sa27 plasmid pSa27-TC-CIP (coverage 100%, e-value 1e-56)
Enterobacter hormaechei strain C15117 plasmid pSPRC-Echo1, (coverage 100%, e-value 1e-56).
May I ask if IGV is copying the sequence of the reads (as a consensus) or that of the reference genome? Since the reference does not have bacterial sequences, how could BLAST find bacteria instead? Would it be because the BLAST algorithm has missed the hit? Or the reads are not really mapped to their expected loci?
Thank you


Pure speculation.
Genome V(since you wish to keep it secret) could have some contamination (or just a region that happens to be similar to a similar sequence in bacteria). If you omit bacteria what else does it hit viablast?Is no secret: V stands for
viral. BLAST gave only bacterial species, but the reference is based only on virus sequences, hence there should be no bacterial hit in the first place. As you pointed out, there might be homology regions, but yet, I was expecting at least a hit on viruses.