Dear Colleagues,
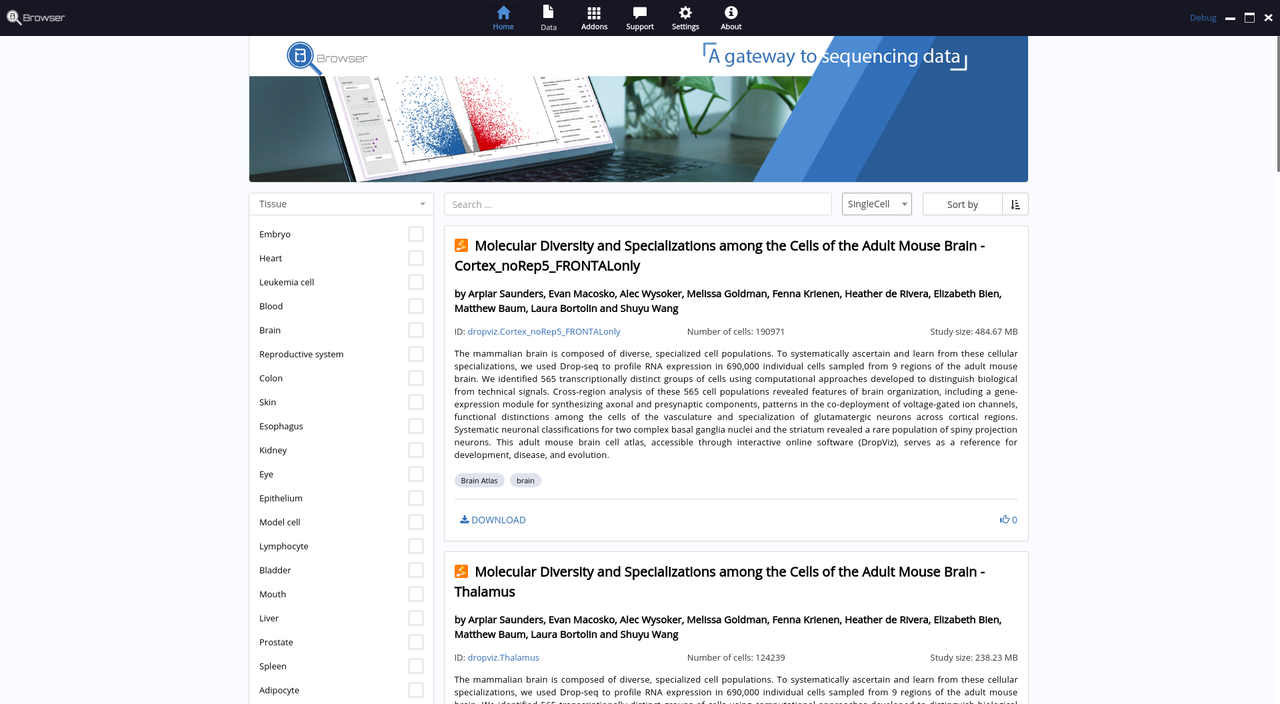
Today, we are happy to release BioTuring Browser, a modern application for accessing published sequencing data, focusing on single-cell transcriptome. With a web browser, scientists can easily read published papers; with BioTuring Browser, they can actually “read” the underneath sequencing data, and answers many questions that were not revealed in the papers.
In this beta release, BioTuring Browser provides:
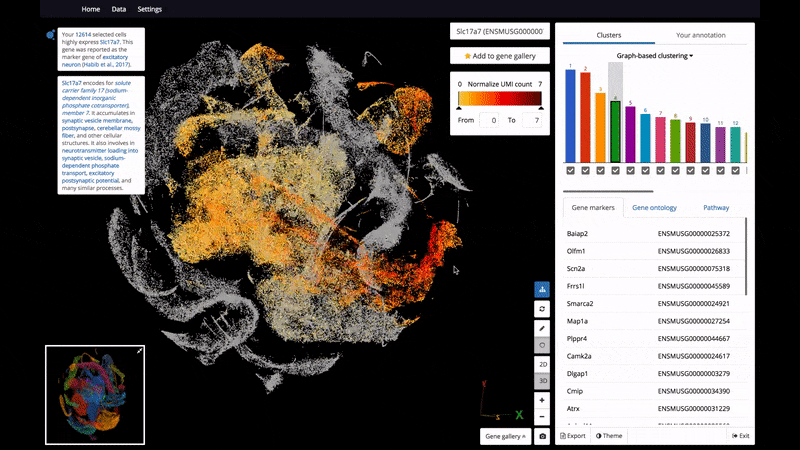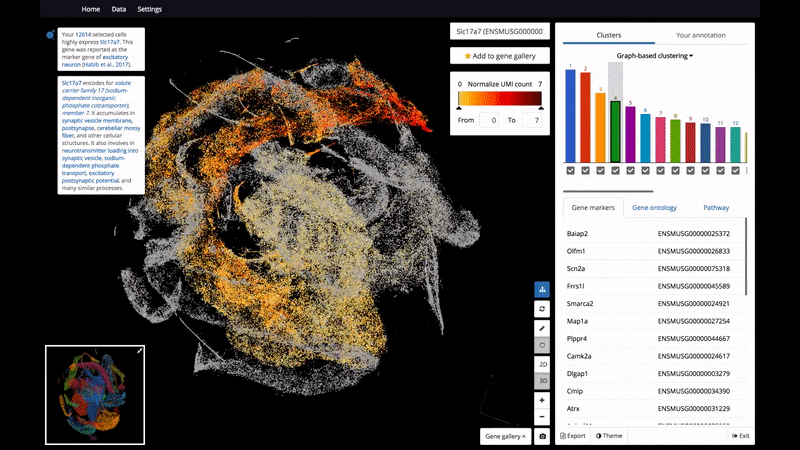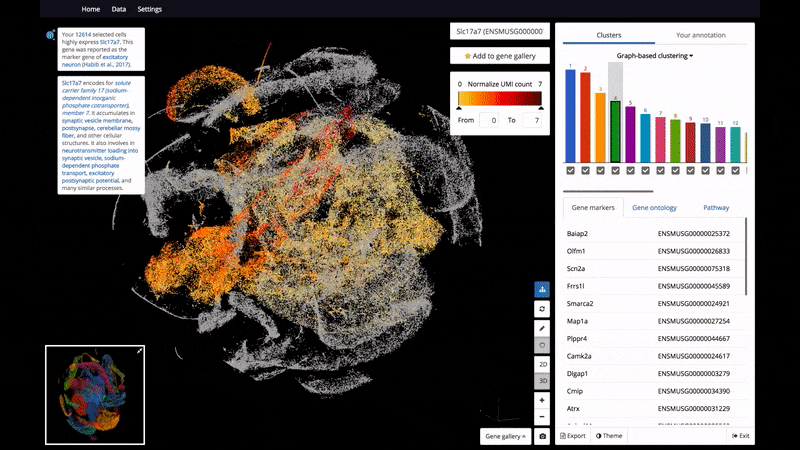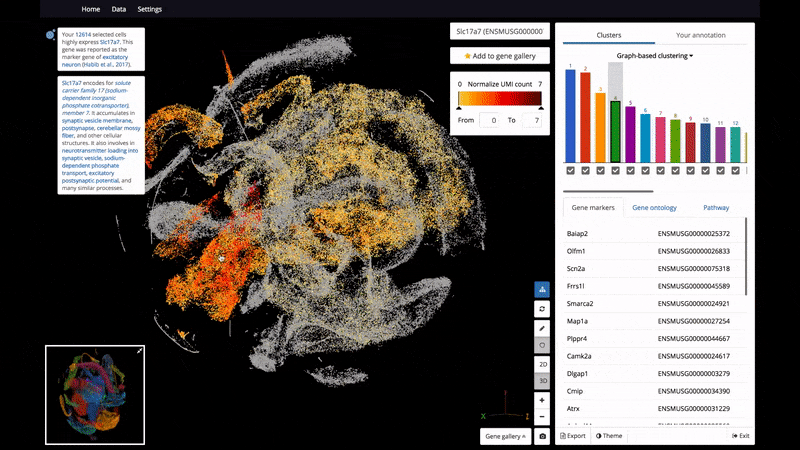
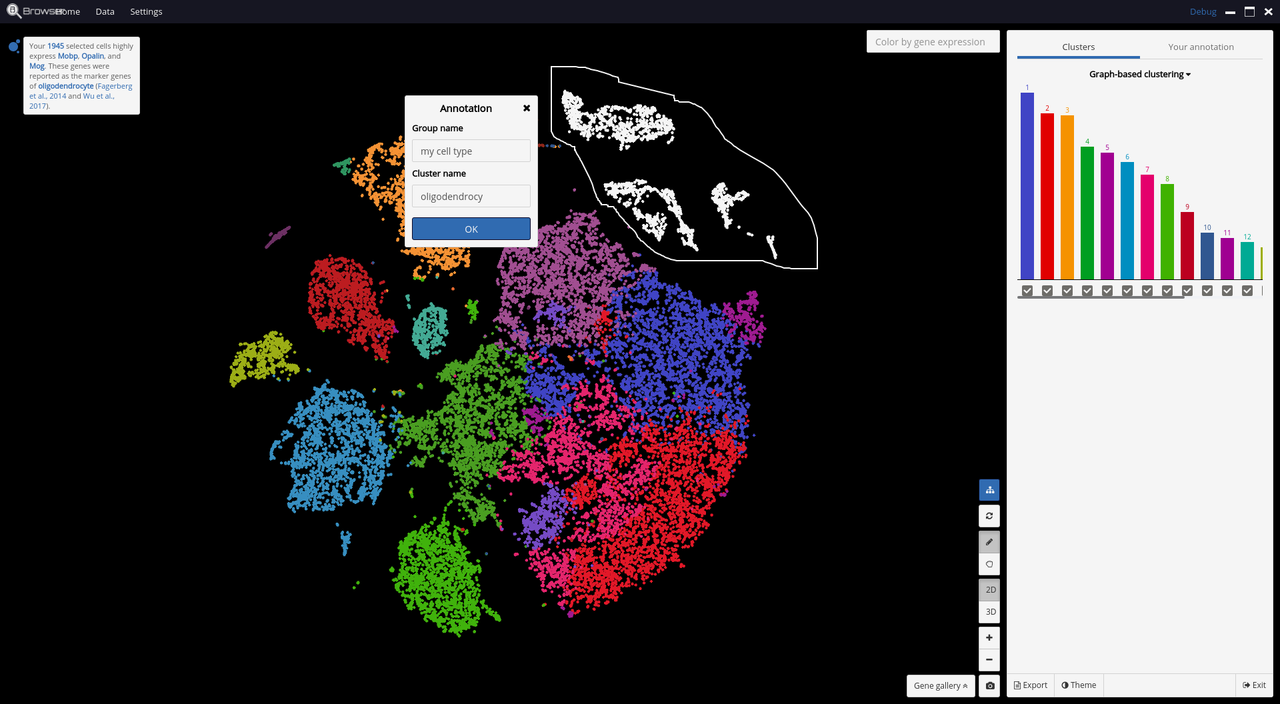
- 3D and 2D PCA/t-SNE visualization of single cells, scaled up to 1.3 million cells on a standard laptop.
- Batch effect removal
- Comparison of two conditions (Differentially Expressed Genes and Comparing Cluster Structure)
- Support automatic and manual cell-type annotations
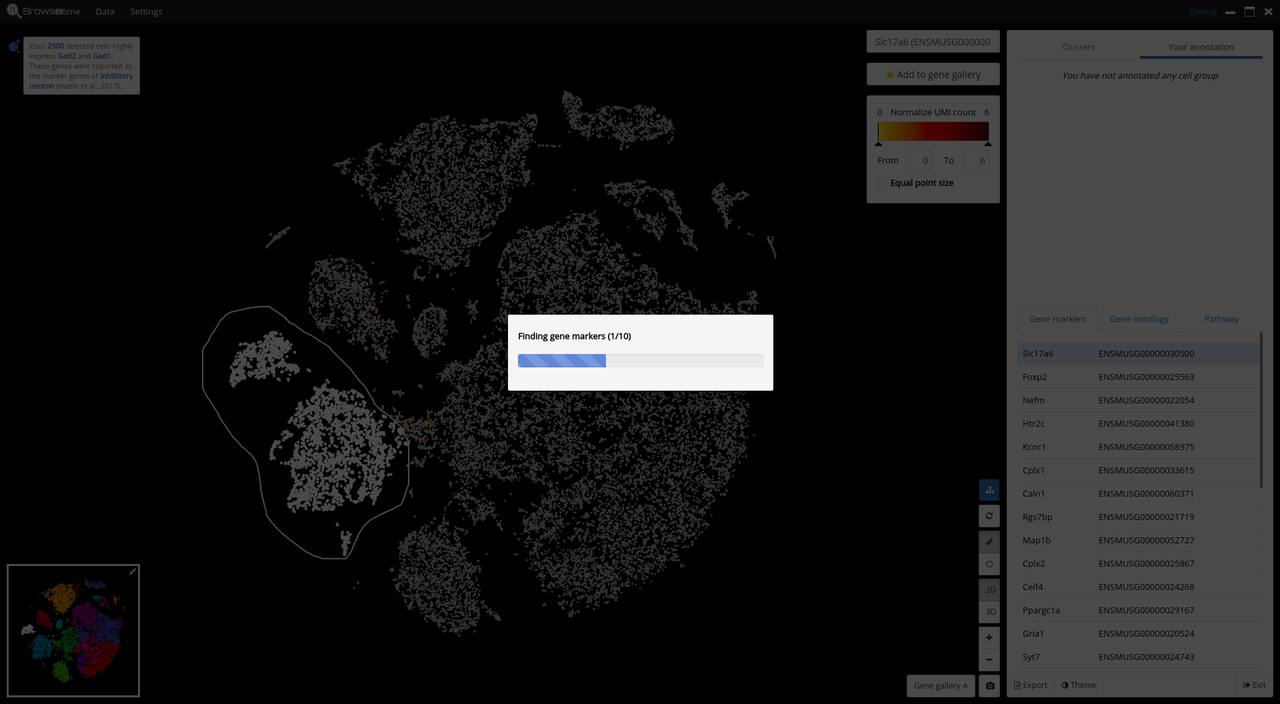
- Support real-time cell type prediction
- Support color mapping for graph-based clustering, k-means clustering, and gene expression.
- Sub-clustering of a chosen cell population.
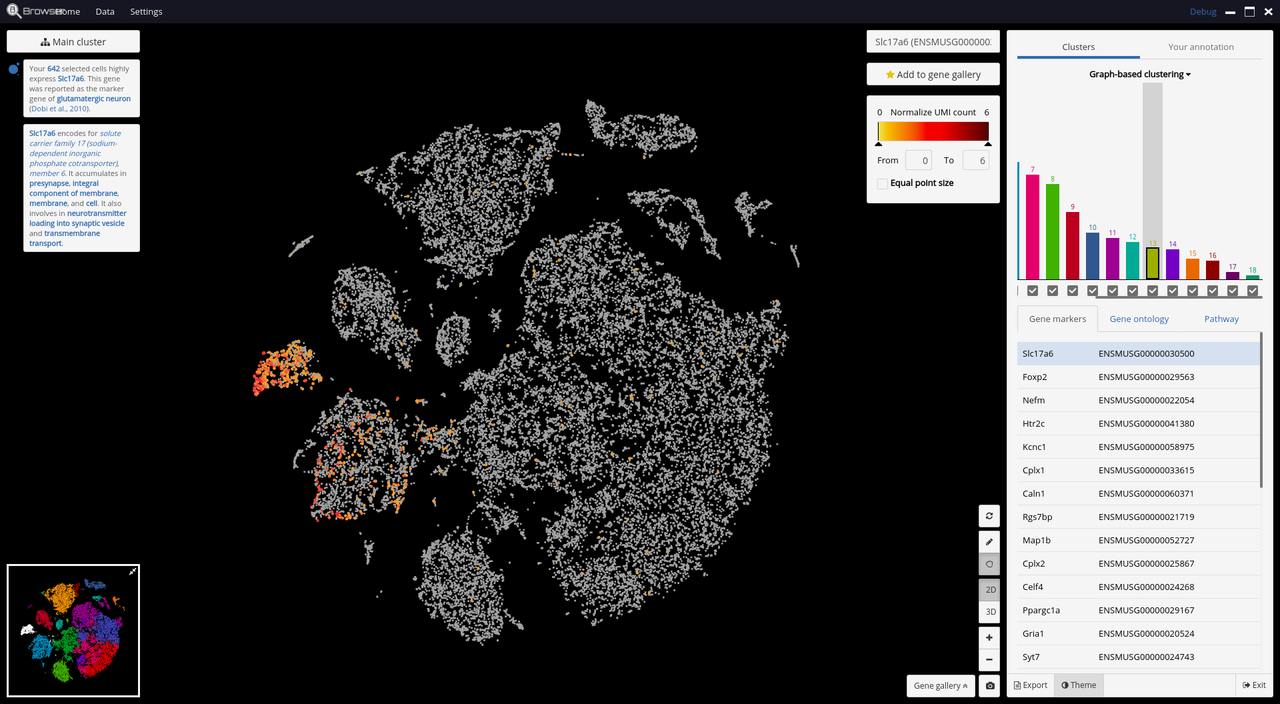
- Marker gene identification
- A comprehensive knowledge base for gene functions.
- Access data from new and highly cited single-cell papers in neuroscience
- Also, the Bulk-RNA seq add-on provides all standard analyses, e.g., PCA, differential expression, gene ontology, pathway enrichment, tissue prediction, etc.
You can download BioTuring Browser at https://bioturing.com/product/bbrowser
Currently, BioTuring Browser is available for MacOS and Windows (beta). We hope to have the Linux version available soon.
Thank you!
Son Pham & BioTuring Team.
Below are some screenshots/videos of BioTuring Browser


Great work! Is it commercial?
You can view published data free of charge. For analyzing private data, we charge by each add-on.
Is there a documentation that explicitely explains all relevant steps, such as filtering criteria, data normalization etc?
Hi, I think this would help: https://static.bioturing.com/v20181025/pdf/BBrowser_Documentation.pdf
I've got 2 batches to handle. How to remove the batch effects with this tool? Can you share some instructions?