Hi All,
I have been trying to combine all genomic resources produced by different sequencing platforms in our lab and assemble them into contigs.
Since we do not have reference genome for our species, we did de novo assembly.
When we finish the assembly, we still have a lot of un-used reads (~30GB).
It doesn't seem to be very reasonable to simply discard them. But i do not know what should i do to take advantage of them.
I am wondering if anyone of you has similar experience and know how to do with it.
Thanks in advance for your valuable suggestions and discussions!

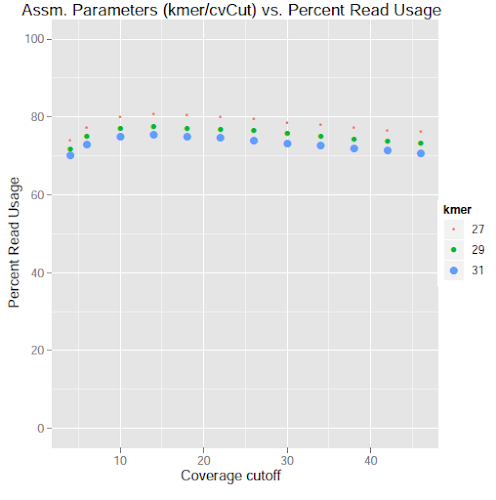

thanks so much for offering this many possibilities. In fact, i have already filtered out potential contaminations. And before assembly i did quality check and filtering. So i think, as you pointed out the effects of lower coverage and repetition could be the reason. I will check that. Many thanks Michael.