I'm very new to this space, but from what I've read I'm not sure I understand what the process of variant calling looks like -- if we're given a reference genome, and then a specific genome of interest, what makes finding variants, which are just differences from the reference, so hard to identify?
For example, at a specific location the sequence, why is it wrong to simply take the majority of whatever bp is there and say "at this position, we are __% confident that individual X has this bp"? And since you would expect see heterozygosity at many loci, what does it even mean if you see "60% A and 40% T" at a given location?
It seems like alignment would be the actual tricky part of the process, not the 'calling' part.
I think i'm probably making some faulty assumptions or do not fully understand the problem, and I'd appreciate any explanation.

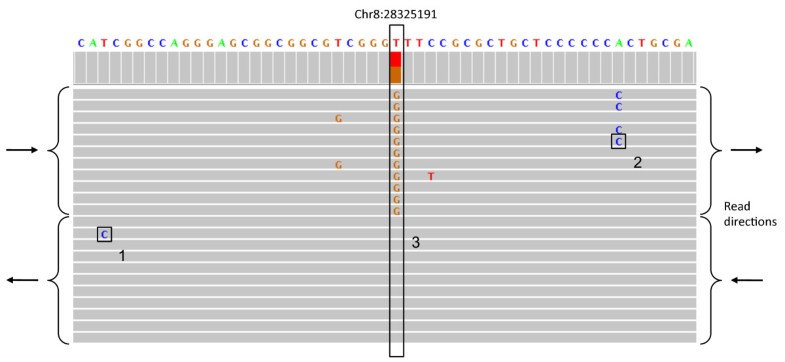

It's worth noting that this inherent representational ambiguity is also why _matching_ variants (for example looking up called variants in a variant database, intersecting two variant call sets to find common vs different variants, evaluating variant caller accuracy with respect to a gold standard benchmark dataset such as GIAB) is not as straight forward as people think. The most accurate variant comparisons require haplotype awareness (e.g. using a tool such as rtg vcfeval or hap.py).
Even something as apparently simple as classifying a variant is a SNP vs Indel can be confounded by this, see the example under "Benchmarking performance for SNPs versus indels" in the vcfeval manual.