Hi, I'm not sure if there is a reason to worry, but i would like to try and understand the problem.
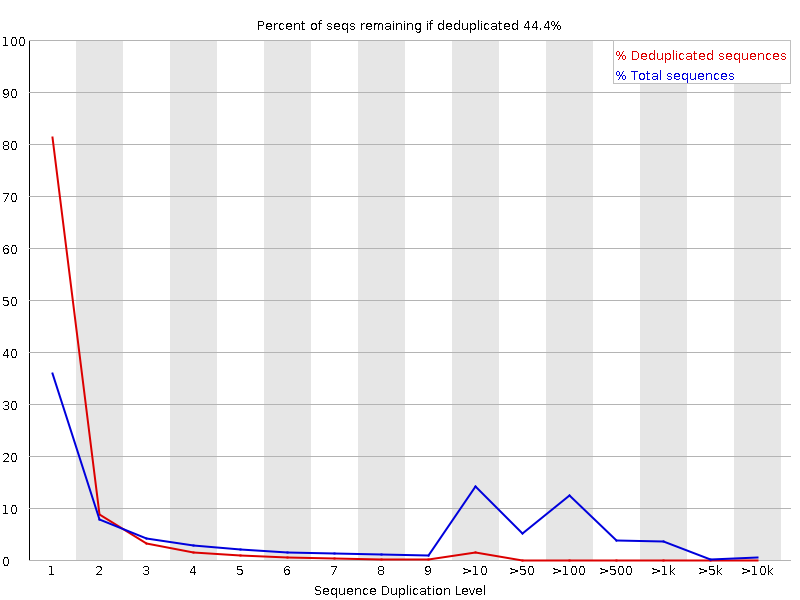
we have a 10x Genomics run sequenced in a nextSeq 500. I know that during the sequencing there can be duplications, but for the first time, we're now seeing that in the R1 reads, so basically, where the 10x-barcode and the UNI-barcode are sequenced.
in the run we have almost 40 samples, but not all of them are showing this behavior. I'm attaching the images of the two samples. R2 duplication is normal, and what it always looks like, but R1 is strange.
Has anyone seen this before and maybe has an explanation for that?
thanks
R1
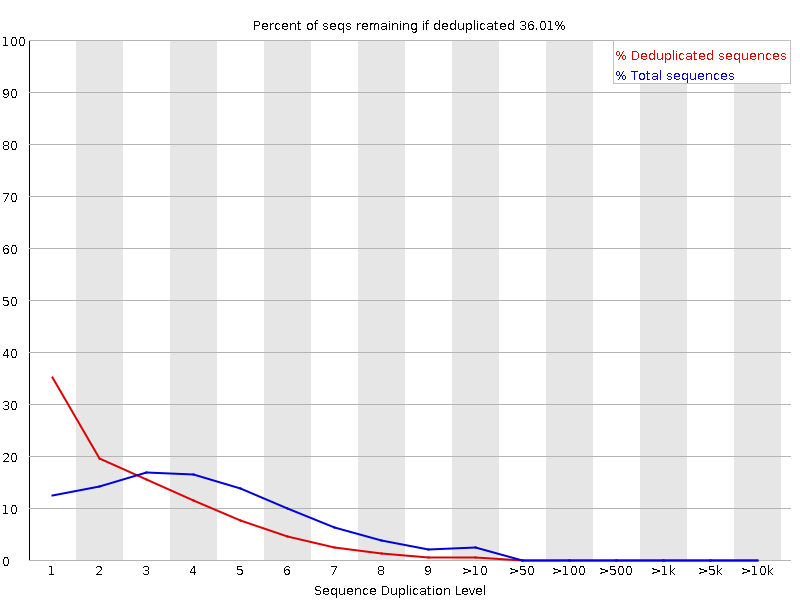
R2